 |
 |
- Search
| Psychiatry Investig > Volume 8(2); 2011 > Article |
Abstract
Objective
To analyze both differentially expressed genes and the Bcl-xL protein expression after acute and chronic treatment with fluoxetine in rat C6 glioma cells.
Methods
C6 glioma cells were cultured for 24 h or 72 h after treatment with 10 ┬ĄM fluoxetine, and gene expression patterns were observed using microarray and qRT-PCR. Then, cells were cultured for 6 h, 24 h, 72 h or 96 h after treatment with 10 ┬ĄM fluoxetine, and the expression of Bcl-xL protein was measured using western blot.
Results
As determined by microarray, treatment with fluoxetine for 24 h up-regulated 33 genes (including Bcl-xL and NCAM140) and down-regulated 7 genes (including cyclin G-associated kinase). Treatment with fluoxetine for 72 h up-regulated 53 genes (including Gs╬▒ and Bcl-xL) and down-regulated 77 genes (including G╬▒i2 and annexin V). Based on the qRT-PCR results, there was an increase in Gs╬▒ mRNA and a decrease in G╬▒i2 mRNA at 72 h in fluoxetine-treated cells as compared to control, a result that was consistent with microarray. We also observed an increase in Bcl-xL mRNA (both at 24 h and at 72 h) in fluoxetine-treated cells as compared to control, demonstrating a tendency to increase gradually. Bcl-xL protein expression increased as the duration of fluoxetine treatment increased.
Fluoxetine, one of the selective serotonin reuptake inhibitors, shows a strong antidepressive effect and is widely used to augment the actions of serotonin (5-HT) in the nervous system.1,2 However, like other antidepressants, the clinical effects of fluoxetine do not begin to appear until a few weeks after the patient has begun taking the drug.3
Previous studies have shown that chronic treatment with antidepressants, including fluoxetine and tianeptine, regulates the expression of genes related to intracellular signal transduction pathways, including pathways involving cyclic adenosine monophosphate (cAMP) responsive element binding protein (CREB), brain-derived neurotropic factor (BDNF), mitogen-activated protein kinase and cell survival, indicating that antidepressants may keep nerve cells from being damaged and also help them regenerate.4-7 Nibuya et al.4 demonstrated that chronic treatment with antidepressants, especially imipramine and fluoxetine, increased CREB and BDNF expression mRNA in rat hippocampus, whereas Reagen et al.6 found that tianeptine increased BDNF protein levels, not in the rat hippocampus (as expected), but in the rat amygdala. This result demonstrated that the brain regions that are targeted by antidepressants can differ. Recently, chronic treatment (8 weeks) of fluoxetine and amitriptyline was shown to increase cyclic guanosine monophosphate but not cAMP in rat hippocampus, suggesting that sustained antidepressant treatment may alter cyclic mononucleotide messenger systems.7 However, it is still not fully known how the expression of genes related to the cellular mechanisms responsible for the therapeutic effect of antidepressants changes during treatment.
In a previous study, we identified genes that may be involved in the therapeutic response of fluoxetine using microarray technology, which can simultaneously monitor the expression levels of thousands of genes.8 In our study using rat basophilic leukemia (RBL-2H3) cells, fluoxetine mediated the up- and down-regulation of several genes, including 14-3-3 eta, which was down-regulated at 24 h and 36 h and then up-regulated at 48 h. Even though our previous research described a new method for analyzing the expression pattern of several genes that are simultaneously regulated by fluoxetine, there were a few limitations (such as the use of cells that did not originate in the brain and the lack of a chronic treatment paradigm for fluoxetine).
The rat C6 glioma cells used in the present study are suitable for studying the effects of antidepressants in vitro because the cells lacking presynaptic input exhibit directly monoamine receptor downregulation through postsynaptic action of the antidepressants.9 In addition, C6 glioma cells have N-methyl-D-aspartate (NMDA) and dopamine D2 receptors as previously mentioned.10,11 Previous studies have observed 5-HT receptor-mediated effects using C6 glioma cells.12,13 Therefore, we can observe a direct postsynaptic effect as well as the biological functions mediated by NMDA, D2 and 5-HT receptors in the cells after fluoxetine treatment. In the present study, we analyzed the genes that were up- and down-regulated by acute (24 h) and chronic (72 h) treatment with fluoxetine in the cells using cDNA microarray. We then confirmed the changes in Gs╬▒, G╬▒i2, and Bcl-xL genes, which were seen in the microarray data, using quantitative RT-PCR (qRT-PCR). In addition, we measured the expression level of Bcl-xL protein after fluoxetine treatment using western blot.
The rat C6 glioma cells (KCLB No;10107) were purchased from KTCC (Seoul, Korea), a rat brain 10K cDNA chip was obtained from Gaiagene (Seoul, Korea) and iQ SYBR green supermix was purchased from Bio-Rad (Hercules, CA, USA). BCL-xL antibody was purchased from Cell Signaling Technology (Beverly, MA, USA) and ╬▓-actin was obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Fluoxetine was provided by Lilly Korea, Ltd. (Seoul, Korea).
C6 cells were grown overnight in Dulbecco's modified eagle medium containing L-glutamate, 4.5 g/L of glucose, 10% fetal bovine serum, 100 units of penicillin, and 100 ┬Ąg/mL of streptomycin at 37Ōäā in humidified 5% CO2. As a control, the cells were fed with fresh media and cultured for 24 h and 72 h. For the fluoxetine treatment group, the cells were fed with fresh media containing 10 ┬ĄM of fluoxetine and cultured for 24 h and 72 h, as was done in a previous study.14
cDNA microarray analysis was performed using a chip containing 2,500 ESTs and 2,500 known genes. The total RNA from the control and fluoxetine-treated cells was isolated using TRIzol┬« Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Fluorescent (Cy3, Cy5)-labeled cDNA probes were made from 20 ┬Ąg of total RNA from both the control cells and the fluoxetine-treated cells, using an amino-allyl cDNA labeling kit (Ambion, Austin, TX, USA). For each experiment, at least four replicates were performed, and two of these replicates were repeated with the fluorophores reversed to eliminate false-positive results. Cy3- and Cy5-labeled probes were mixed with water, 8 ┬Ąg of poly (dA) (Pharmacia), 4 ┬Ąg of Escherichia coli tRNA (Sigma-Aldrich, Saint Louis, MO, USA), 10 ┬Ąg of mouse Cot1 DNA (Invitrogen, Carlsbad, CA, USA) and 60 ┬ĄL of 2X EasyHyb buffer (U-vision Biotech Inc., Taipei, Taiwan) to a final volume of 120 ┬ĄL. The resulting mixture was incubated at 95Ōäā for 5 min. Probes were then applied to the array for hybridization at 52Ōäā for 8 h, using a GeneTAG automatic hybridizer (Genomic Solutions, Boston, MA, USA). After hybridization, the slides were washed first in 2├ŚSSC and 0.2% SDS at 52Ōäā for 5 min, then with 0.1├ŚSSC and 0.2% SDS at 52Ōäā for 5 min and finally with 0.1├ŚSSC for 5 min at room temperature. The slides were dried by spinning for 3 min. The dried slides were scanned with a ScanArray 5,000 fluorescence reader, and QuantArray image acquisition software (PerkinElmer Life Sciences) was used to quantify the signal and background intensity for each target element. The ratio of the two corrected signal intensities was calculated and used as the differential expression ratio (DE) for each specific gene in the two mRNA samples. The raw data of the repeated experiments were converted to log ratios. Negative control spots, small signal-to-noise ratio spots that were less than 2 and spots whose average sensitivity was less than 7.5 were removed, and the data were standardized by Lowess smoothing. Four repeated experiments were standardized by the significant analysis of microarray (SAM; t-statistics) method. Genes differentially expressed by fluoxetine were linkage-analyzed using Cluster 3.0 (open source clustering software) and visualized using JavaTreeview-1.1.3 software (Human Genome Center, Institute of Medical Science, Tokyo, Japan).
Total RNA from fluoxetine-treated and control cells was isolated using TRIzol┬« Reagent and reverse transcribed to cDNA in order to confirm the expression patterns of the genes Gs╬▒, G╬▒i2, and Bcl-xL which were differentially expressed by fluoxetine treatment on cDNA microarray result. For qRT-PCR, 12.5 ┬ĄL of SYBR┬« Premix Ex TaqTM II (Takara BIO, Shiga, Japan), 0.5 ┬ĄL of ROX, 5 pmol of forward primer, 5 pmol of reverse primer and 0.5 ┬ĄL of cDNA were mixed with water to a final volume of 25 ┬ĄL. The mixture was amplified for 40 cycles with an ABI 7500 Real-Time PCR System (Applied Biosystems, Inc., Foster City, CA) with an initial melt at 95Ōäā for ten min followed by 40 cycles of 95Ōäā for 15 sec and 60Ōäā for one min. The cycle number at which a statistically significant increase in each gene was first detected (threshold cycle, Ct) was then normalized to the Ct for ╬▓-actin, which was used as an internal control. The relative expression differences between the control and fluoxetine-treated cells were calculated using the 2-╬ö╬öCT method.15 The primers used to amplify the described genes were listed in Table 1. Primers were designed with Genamics Expression software (Genamics, Hamilton, New Zealand).
The fluoxetine-treated cells were cultured in media containing 10 ┬ĄM of fluoxetine for 6 h, 24 h, 72 h or 96 h. The cells were then washed in phosphate-buffered saline and harvested. Total protein was extracted from the cells using Pro-PrepTM (iNtRON Biotechnology, Korea) according to the manufacturer's instructions. The protein was quantified using the Bradford method (Bio-Rad Protein Assay Kit; Bio-Rad, Hercules, CA, USA). The protein was separated on 12% polyacrylamide gel through sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrotransferred onto a nitrocellulose membrane (Bio-Rad, Hercules, CA, USA). After blocking with 5% non-fat dry milk in TBST (20 mM Tris-HCl, pH 7.6; 500 mM NaCl, 0.1% Tween 20), the membrane was incubated overnight at 4Ōäā with the polyclonal rabbit anti-Bcl-xL antibody (1 : 1,000), followed by incubation with HRP-conjugated anti-mouse IgG (diluted 1 : 1,000) for 60 min. After washing in TBST, the reactions were detected using the enhanced chemiluminescence detection system (Amersham Biosciences, Sunnyvale, CA, USA). The concentration of Bcl-xL exposed on X-ray film was quantified with QuantityOne software (Bio-Rad, USA).
Data were expressed as the mean┬▒standard error. A nonlinear-fitting program (OriginLab Corp., Northampton, MA, USA) was used. Paired and unpaired Student's t-tests were used as appropriate to evaluate the statistical significance of differences between the two groups (fluoxetine-treated and control groups). Values of p<0.05 were considered to indicate statistical significance.
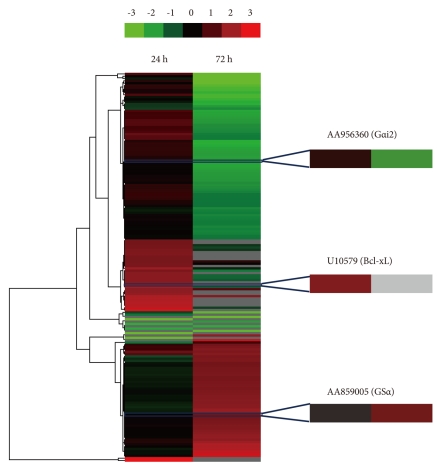
The gene expression profile of control and 10 ┬ĄM fluoxetine-treated cells after 24 h and 72 h was analyzed with a chip containing 2,500 ESTs and 2,500 known genes. After mining biological data using log_2 (a fold change cut-off of 1.5), a hierarchical dendrogram of 169 genes was completed by clustering differentially expressed genes at 24 h and 72 h after fluoxetine treatment (Figure 1). Of the 2,500 known genes in the chip, 7 genes were down-regulated more than 1.4-fold in cells treated with fluoxetine for 24 h compared to the control cells, and 77 of 2,500 known genes were down-regulated after 72 h of fluoxetine treatment. Meanwhile, 33 genes were up-regulated more than 1.4-fold in fluoxetine-treated cells after 24 h compared to control cells, and 53 genes were up-regulated more than 1.4-fold in fluoxetine-treated cells after 72 h of treatment.
Table 2 shows a summarized list of time-dependently expressed genes between the control and fluoxetine-treated C6 cells. Of the down-regulated genes in fluoxetine-treated cells at 24 h, cyclin G-associated kinase, which assists heat shock cognate 70 in uncoating clathrin-coated vesicles16,17 decreased in expression by two-fold. In the fluoxetine-treated cells at 24 h, Bcl-xL and neural cell adhesion molecule 140 (NCAM140) were up-regulated. Bcl-xL (one of the Bcl-2 family members) is widely expressed in the adult brain, despite the relative lack of physiological apoptosis.18
Annexin V and G╬▒i2 were down-regulated in fluoxetine-treated cells at 72 h, whereas 14-3-3 eta and Gs╬▒ were up-regulated. Compared to the expression changes at 24 h, more genes showed a change in expression at 72 h.
To confirm the result of the microarray analysis, we performed qRT-PCR on RNA isolated from control and fluoxetine-treated cells. Gs╬▒ mRNA expression was 1.03┬▒0.12-fold greater at 24 h and 2.63┬▒0.24-fold greater at 72 h in fluoxetine-treated cells compared to the control cells as observed in our previous study (Figure 2A).15 On the other hand, G╬▒i2 expression was 0.86┬▒0.06-fold at 24 h and 0.72┬▒0.05-fold at 72 h compared to control as observed in our previous study (Figure 2B).15 Gene expression of Gs╬▒ and G╬▒i2 at 24 h was not significantly different from control levels. However, at 72 h, gene expression of Gs╬▒ and G╬▒i2 was significantly different from expression in the control cells. Chronic treatment of fluoxetine induced up-regulation of Gs╬▒ and down-regulation of Gi╬▒2, which was also found in a previous study.19 Bcl-xL mRNA significantly increased by 1.36┬▒0.13-fold at 24 h and by 1.65┬▒0.13-fold at 72 h in fluoxetine-treated cells as compared to control cells (Figure 2C). The expression in all three of these genes changed more after chronic treatment than after acute treatment.
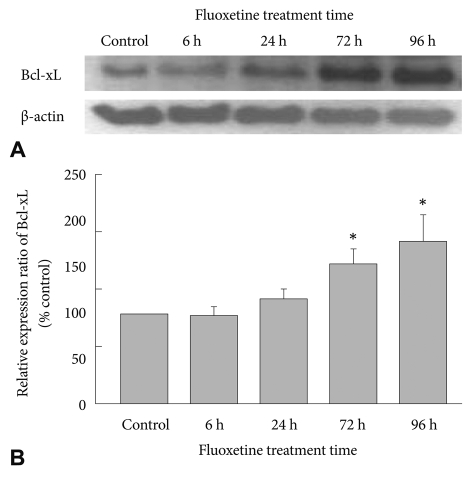
After treating C6 cells with 10 ┬ĄM fluoxetine and culturing them for 6 h, 24 h, 72 h or 96 h, we observed the expression pattern of Bcl-xL protein. Expression gradually increased as treatment time progressed (Figure 3A). The increase of Bcl-xL expression at 6 h and 24 h was not significantly different in fluoxetine-treated cells compared to the control cells (Figure 3B). However, at 72 h and 96 h, there was a significant increase in Bcl-xL expression compared to control. Therefore, fluoxetine increased the expression of the Bcl-xL protein over time.
Over the past few decades, theories on depression have suggested that atrophy of neurons from stress, apoptosis or depression may result from an impairment of the intracellular signaling pathway and a failure of neurons to appropriately adapt.20,21 Studies on antidepressants and depression have mainly focused on the molecular and cellular antidepressant-induced changes that lead to neurogenesis and restored synaptic activity.
We analyzed the genes that were up- and down-regulated as a result of acute and chronic treatment with fluoxetine in C6 cells and detected expression changes in genes that engage in the intracellular signal transduction cascade, including the following: Gs╬▒ and G╬▒i2; neurite outgrowth, including NCAM140; and cell survival, such as annexin V and Bcl-xL. Depressed patients exhibit dysfunction of the cAMP signaling pathway, namely a decrease in adenylyl cyclases, the enzymes that generate intracellular cAMP.22 This dysfunction occurs in the cerebral cortices of patients with depression. Adenylyl cyclase is known to bind with the stimulatory G protein (Gs) and subsequently increase cAMP and cAMP-dependent protein kinase.23 In the present study, we observed that chronic treatment with fluoxetine induced expression of Gs╬▒ (the stimulatory ╬▒-subunit of the Gs protein) mRNA more than 2.5-fold compared to untreated control cells, a finding that was similar to previous studies.10,24 Toki et al.24 demonstrated that chronic treatment of C6 glioma cells with fluoxetine induced a 2-fold increase of Gs╬▒ in the plasma membrane compared to control cells and also promoted the binding of Gs╬▒ and adenylyl cyclase. Chen and Rasenick10 reported that chronic treatment with desipramine, one of the tricyclic antidepressants, enhanced the binding of Gs╬▒ and adenylyl cyclase by 1.7-fold compared to control. In the present study, G╬▒i2 (an antagonist of Gs╬▒) mRNA expression was seldom different in cells with acute treatment of fluoxetine compared to control cells, but chronic treatment decreased its expression by 2-fold in the fluoxetine-treated cells. In a study by Emamghoreishi et al.,19 chronic treatment with desipramine induced a 10% decrease of G╬▒i2 expression in rat cerebral cortex, but the decrease was not significantly different from expression in the control group. When considering the previous studies10,19,24 along with our work, chronic treatment with antidepressants (specifically fluoxetine) can clearly lead to an increase of Gs╬▒ and a decrease of G╬▒i2, suggesting that fluoxetine may activate the cAMP signaling pathway. This activation induces genes related to neuronal survival and synaptic remodeling.
NCAM, which has three major isoforms (NCAM180, 140 and 120), regulates structural and synaptic plasticity in the brain.25,26 In particular, NCAM140 promotes neurite outgrowth.27 A recent report showed that NCAM140 mRNA is decreased in the peripheral blood cells of patients with bipolar disorder, indicating that this change may be associated with the pathophysiology of the disease.28 Interestingly, we found that NCAM140 mRNA expression was increased after acute fluoxetine treatment. We hypothesize that fluoxetine may promote neurite outgrowth of damaged neural cells in the brains of depressed patients through up-regulation of NCAM140.
Annexin V is a Ca2+-dependent protein that combines with phospholipids phosphatidylserine (PS) after the translocation of PS from the inside to the outside of the plasma membrane. Annexin V expression increases in the early stages of cell death.29 In the present study, we observed that chronic treatment with fluoxetine led to a 2-fold down-regulation of annexin V compared to control cells.
In the present study, Bcl-xL expression (both mRNA and protein) in the C6 cells was up-regulated by both acute and chronic treatment with fluoxetine, revealing a tendency for the expression level of Bcl-xL to rise as the duration of fluoxetine treatment grew longer. As already mentioned, neurotrophic factors (such as CREB and BDNF) are regulated by stress and antidepressants through cell survival pathways that also control the Bcl-2 family proteins.30,31 Of the Bcl-2 family, Bcl-2 and Bcl-xL are localized to the mitochondrial outer membrane, act as antiapoptotic proteins in intracellular apoptotic signal transduction32 and are up-regulated by treatment with antidepressants.5,33 Kosten et al.33 used an animal model of depression to show that both repeated stress and the chronic administration of antidepressants (such as fluoxetine and reboxetine) differentially regulated Bcl-xL mRNA expression in the rat hippocampus. They reported that reboxetine increased Bcl-xL mRNA in the hippocampus but that fluoxetine did not have an effect on Bcl-xL. On the other hand, Chiou et al.5 observed that Bcl-xL and Bcl-2 mRNA expression was up-regulated by chronic treatment with fluoxetine in lipopolysaccharide (LPS)-treated neural stem cells (NSCs) derived from the hippocampus; this up-regulation repressed LPS-induced apoptosis in NSCs. Based on the results of the present study as well as previous studies,5,33 we suggest that when fluoxetine regulates the expression of antiapoptotic agents (Bcl-2 and Bcl-xL), the effects are different in vivo and in vitro; for example, Bcl-2 is up-regulated by fluoxetine in vivo, whereas Bcl-xL is mainly up-regulated in vitro. Furthermore, the increase that we see in Bcl-xL protein levels (as well as in mRNA levels) induced by chronic treatment with fluoxetine clearly indicates that fluoxetine inhibits apoptosis through the Bcl-xL-related antiapoptotic pathway. This result is unlike previous studies, which only demonstrated an increase in Bcl-xL mRNA.5,33
In conclusion, our data not only confirmed that chronic treatment of fluoxetine induces Gs╬▒ and repressed G╬▒i2 expression but also demonstrated that expression of Bcl-xL (both mRNA and protein) is increased by chronic treatment with fluoxetine in C6 cells. These results suggest that these genes may play a critical role in delaying the clinical effects of antidepressants. However, additional studies are necessary to further analyze the other genes found in the microarray and their roles in the effects of antidepressants.
References
1. Fuller RW, Wong DT, Robertson DW. Fluoxetine, a selective inhibitor of serotonin uptake. Med Res Rev 1991;11:17-34. PMID: 1994152.


2. Wong DT, Bymaster FP, Engleman EA. Prozac (fluoxetine, Lilly 110140), the first selective serotonin uptake inhibitor and an antidepressant drug: twenty years since its first publication. Life Sci 1995;57:411-441. PMID: 7623609.


3. Vaswani M, Linda FK, Ramesh S. Role of selective serotonin reuptake inhibitors in psychiatric disorders: a comprehensive review. Prog Neuropsychopharmacol Biol Psychiatry 2003;27:85-102. PMID: 12551730.


4. Nibuya M, Nestler EJ, Duman RS. Chronic antidepressant administration increases the expression of cAMP response element binding protein (CREB) in rat hippocampus. J Neurosci 1996;16:2365-2372. PMID: 8601816.



5. Chiou SH, Chen SJ, Peng CH, Chang YL, Ku HH, Hsu WM, et al. Fluoxetine up-regulates expression of cellular FLICE-inhibitory protein and inhibits LPS-induced apoptosis in hippocampus-derived neural stem cell. Biochem Biophys Res Commun 2006;343:391-400. PMID: 16545775.


6. Reagan LP, Hendry RM, Reznikov LR, Piroli GG, Wood GE, McEwen BS, et al. Tianeptine increases brain-derived neurotrophic factor expression in the rat amygdala. Eur J Pharmacol 2007;565:68-75. PMID: 17368617.


7. Reierson GW, Mastronardi CA, Licinio J, Wong ML. Repeated antidepressant therapy increases cyclic GMP signaling in rat hippocampus. Neurosci Lett 2009;466:149-153. PMID: 19788915.



8. Baik SY, Jung KH, Choi MR, Yang BH, Kim SH, Lee JS, et al. Fluoxetine-induced up-regulation of 14-3-3zeta and tryptophan hydroxylase levels in RBL-2H3 cells. Neurosci Lett 2005;374:53-57. PMID: 15631896.


9. P├Īv M, Kov├Īr┼» H, Fiserov├Ī A, Havrdov├Ī E, Lis├Ī V. Neurobiological aspects of depressive disorder and antidepressant treatment: role of glia. Physiol Res 2008;57:151-164. PMID: 17465696.

10. Chen J, Rasenick MM. Chronic treatment of C6 glioma cells with antidepressant drugs increases functional coupling between a G protein (Gs) and adenylyl cyclase. J Neurochem 1995;64:724-732. PMID: 7830066.


11. Roh K, Roh S, Yang BH, Lee JS, Chai YG, Choi MR, et al. Effects of haloperidol and risperidone on the expression of heat shock protein 70 in MK-801-treated rat C6 glioma cells. Prog Neuropsychopharmacol Biol Psychiatry 2008;32:1793-1797. PMID: 18721842.


12. Elliott JM, Newberry NR, Cholewinski AJ, Bartrup JT, Briddon SJ, Carey JE, et al. Characterization of the 5-hydroxytryptamine2A receptor-activated cascade in rat C6 glioma cells. Neuroscience 1995;69:1119-1131. PMID: 8848101.


13. Thomas EA, Matli JR, Hu JL, Carson MJ, Sutcliffe JG. Pertussis toxin treatment prevents 5-HT(5a) receptor-mediated inhibition of cyclic AMP accumulation in rat C6 glioma cells. J Neurosci Res 2000;61:75-81. PMID: 10861802.


14. Donati RJ, Thukral C, Rasenick MM. Chronic treatment of C6 glioma cells with antidepressant drugs results in a redistribution of Gsalpha. Mol Pharmacol 2001;59:1426-1432. PMID: 11353802.


15. Choi MR, Baik SY, Jung KH, Chai YG, Kim SH, Roh S, et al. Characterization of differentially expressed genes upon chronic fluoxetine treatment in rat C6 glioma cells. Korean J Psychopharmacol 2004;15:457-467.
16. Greener T, Zhao X, Nojima H, Eisenberg E, Greene LE. Role of cyclin G-associated kinase in uncoating clathrin-coated vesicles from non-neuronal cells. J Biol Chem 2000;275:1365-1370. PMID: 10625686.


17. Umeda A, Meyerholz A, Ungewickell E. Identification of the universal cofactor (auxilin 2) in clathrin coat dissociation. Eur J Cell Biol 2000;79:336-342. PMID: 10887964.


18. Krajewski S, Krajewska M, Shabaik A, Wang HG, Irie S, Fong L, et al. Immunohistochemical analysis of in vivo patterns of Bcl-X expression. Cancer Res 1994;54:5501-5507. PMID: 7923184.

19. Emamghoreishi M, Warsh JJ, Sibony D, Li PP. Lack of effect of chronic antidepressant treatment on Gs and Gi alpha-subunit protein and mRNA levels in the rat cerebral cortex. Neuropsychopharmacology 1996;15:281-287. PMID: 8873111.


20. Duman RS, Malberg J, Thome J. Neural plasticity to stress and antidepressant treatment. Biol Psychiatry 1999;46:1181-1191. PMID: 10560024.


21. McEwen BS. Stress and hippocampal plasticity. Annu Rev Neurosci 1999;22:105-122. PMID: 10202533.


22. Dowlatshahi D, MacQueen GM, Wang JF, Reiach JS, Young LT. G Protein-coupled cyclic AMP signaling in postmortem brain of subjects with mood disorders: effects of diagnosis, suicide, and treatment at the time of death. J Neurochem 1999;73:1121-1126. PMID: 10461903.


23. Ozawa H, Rasenick MM. Chronic electroconvulsive treatment augments coupling of the GTP-binding protein Gs to the catalytic moiety of adenylyl cyclase in a manner similar to that seen with chronic antidepressant drugs. J Neurochem 1991;56:330-338. PMID: 1898967.


24. Toki S, Donati RJ, Rasenick MM. Treatment of C6 glioma cells and rats with antidepressant drugs increases the detergent extraction of G (s alpha) from plasma membrane. J Neurochem 1999;73:1114-1120. PMID: 10461902.


25. Benson DL, Schnapp LM, Shapiro L, Huntley GW. Making memories stick: cell-adhesion molecules in synaptic plasticity. Trends Cell Biol 2000;10:473-482. PMID: 11050419.


26. Sandi C. Stress, cognitive impairment and cell adhesion molecules. Nat Rev Neurosci 2004;5:917-930. PMID: 15550947.


27. Buttner B, Reutter W, Horstkorte R. Cytoplasmic domain of NCAM 180 reduces NCAM-mediated neurite outgrowth. J Neurosci Res 2004;75:854-860. PMID: 14994346.


28. Wakabayashi Y, Uchida S, Funato H, Matsubara T, Watanuki T, Otsuki K, et al. State-dependent changes in the expression levels of NCAM-140 and L1 in the peripheral blood cells of bipolar disorders, but not in the major depressive disorders. Prog Neuropsychopharmacol Biol Psychiatry 2008;32:1199-1205. PMID: 18430502.


29. Meers P, Mealy T. Calcium-dependent annexin V binding to phospholipids: stoichiometry, specificity, and the role of negative charge. Biochemistry 1993;32:11711-11721. PMID: 8218240.


30. Riccio A, Ahn S, Davenport CM, Blendy JA, Ginty DD. Mediation by a CREB family transcription factor of NGF-dependent survival of sympathetic neurons. Science 1999;286:2358-2361. PMID: 10600750.


31. Manji HK, Drevets WC, Charney DS. The cellular neurobiology of depression. Nat Med 2001;7:541-547. PMID: 11329053.


33. Kosten TA, Galloway MP, Duman RS, Russell DS, D'Sa C. Repeated unpredictable stress and antidepressants differentially regulate expression of the bcl-2 family of apoptotic genes in rat cortical, hippocampal, and limbic brain structures. Neuropsychopharmacology 2008;33:1545-1558. PMID: 17700647.


Figure┬Ā1
Microarray analysis of fluoxetine-treated C6 glioma cells. Control and 10 ┬ĄM fluoxetine-treated cells were cultured for 24 h or 72 h. Then, 169 differentially expressed genes in these cells were analyzed using cDNA microarray and clustered using a hierarchical dendrogram. In general, genes in the 72 h group were more up- or down-regulated than genes in the 24 h group. The gray color indicates that signal intensity was not identified. Three genes magnified in the hierarchical dendrogram were selected for confirmation of their mRNA expression using qRT-PCR.
Figure┬Ā2
mRNA expression of Gs╬▒, G╬▒i2, and Bcl-xL. C6 glioma cells were incubated with 10 ┬ĄM fluoxetine for 24 h or 72 h and the resulting mRNA from these cells was analyzed by qRT-PCR. A: mRNA levels of Gs╬▒ were significantly increased in fluoxetine-treated cells after 72 h compared to control cells. B: mRNA levels of G╬▒i2 were significantly decreased in fluoxetine-treated cells after 72 h compared to control cells. C: mRNA levels of Bcl-xL were significantly increased in fluoxetine-treated cells after both 24 h and 72 h compared to control cells. Values are expressed as mean┬▒SEM (n=4). *significantly different from control (p<0.05). The data were analyzed by two sample t-test.

Figure┬Ā3
The effect of fluoxetine on Bcl-xL protein expression. C6 glioma cells were incubated with 10 ┬ĄM fluoxetine for 6 h, 24 h, 72 h or 96 h. A: The result of western blotting using anti-Bcl-xL and anti-╬▓-actin antibodies after separating proteins via 12% SDS-PAGE. B: The relative expression density of Bcl-xL in fluoxetine-treated cells compared to the control. The density of Bcl-xL in fluoxetine-treated cells and control cells was normalized with ╬▓-actin prior to comparison of Bcl-xL density between fluoxetine-treated cells and control cells. Results are shown as the mean┬▒SEM (n=4). *significantly different from control (p<0.05). The data were analyzed by two sample t-test. SDS-PAGE: sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Table┬Ā2
Summarized list of time-dependently expressed genes between control and fluoxetine-treated C6 cells
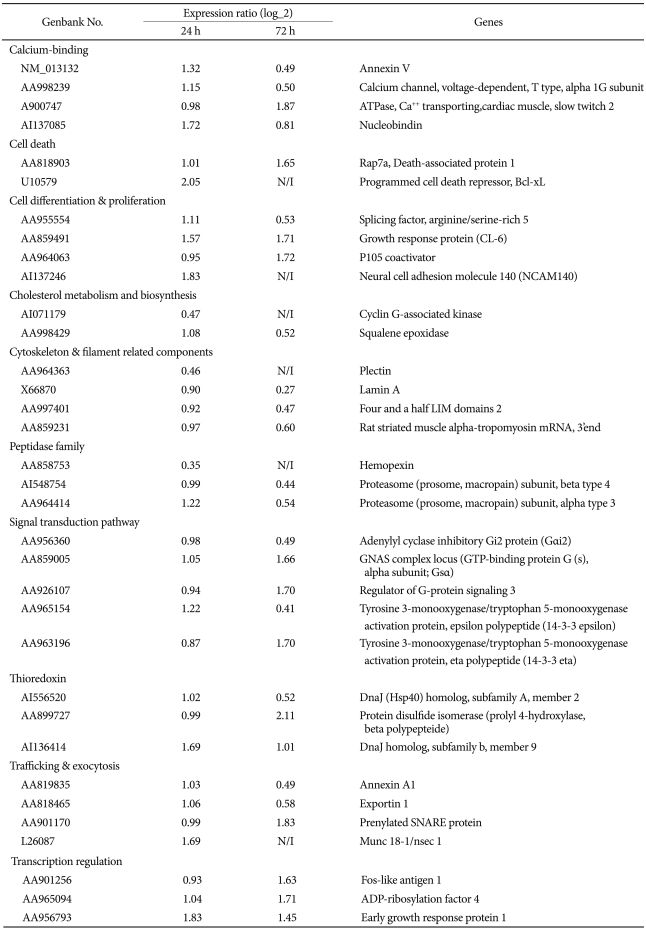
N/I (not identified) shows that signal intensity was not identified. A part of listed genes was already mentioned in our previous study15







