m6A Methyltransferase METTL3 Reduces Hippocampal Neuron Apoptosis in a Mouse Model of Autism Through the MALAT1/SFRP2/Wnt/β-catenin Axis
Article information
Abstract
Objective
Hippocampal neuron apoptosis contributes to autism, while METTL3 has been documented to possess great potentials in neuron apoptosis. Our study probed into the role of METTL3 in neuron apoptosis in autism and to determine the underlying mechanism.
Methods
Bioinformatics analysis was used to analyze expressed genes in autism samples. Institute of Cancer Research mice were treated with valproic acid to develop autism models. The function of METTL3 in autism-like symptoms in mice was analyzed with behavioral tests and histological examination of their hippocampal tissues. Primary mouse hippocampal neurons were extracted for in vitro studies. Downstream factors of METTL3 were explored and validated.
Results
METTL3, MALAT1, and Wnt/β-catenin signaling were downregulated, while SFRP2 was upregulated in the hippocampal tissues of a mouse model of autism. METTL3 stabilized MALAT1 expression by promoting m6A modification of MALAT1. MALAT1 promoted SFRP2 methylation and led to reduced SFRP2 expression by recruiting DNMT1, DNMT3A, and DNMT3B to the promoter region of SFRP2. Furthermore, SFRP2 facilitated activation of the Wnt/β-catenin signaling. By this mechanism, METTL3 suppressed autism-like symptoms and hippocampal neuron apoptosis.
Conclusion
This research suggests that METTL3 can reduce autism-like symptoms and hippocampal neuron apoptosis by regulating the MALAT1/SFRP2/Wnt/β-catenin axis.
INTRODUCTION
Autism is a severe neurobehavioral syndrome and often accompanied with deficient social interaction and communication capacities and repeated and stereotyped behaviors [1]. Autism affects males more than females which might due to the influence of fetal testosterone [2]. More importantly, autism is commonly diagnosed in early childhood [3]. Accumulated evidence reveals that autism is related to aberrant expression of genetic factors or environment factors, including alcohol abuse and cocaine during pregnancy [4]. To get insight to the molecular mechanism, hippocampal neuron apoptosis has been reported to contribute to the development of autism [5].
Recently, m6A modification has been reported to regulate gene expression by affecting mRNA stability, translation, and transcription elongation [6-8]. METTL3, a methyltransferase, governs predominant m6A modification in cells and engages in diverse biological processes, including cell apoptosis [9]. Importantly, a recent study reveals aberrant downregulation of METTL3 in autism hippocampal tissues while the molecular mechanism of how METTL3 regulates the development of autismlike behaviors remains exclusive [10]. METTL3 can mediate the expression of MALAT1 in adriamycin-resistant breast cancer through m6A [11].
Long noncoding RNAs (lncRNAs) are involved in the regulation of various critical biological processes by regulating gene expression, sponging microRNAs or functioning as a scaffold to recruit specific epigenetic factors to the certain sites [12,13]. Bioinformatics analysis predicted that DNA methyltransferase 1 (DNMT1), DNMT3A, and DNMT3B might interact with MALAT1. DNMT protein family has been reported to participate in regulation of biological process by promoting methylation on genes promoter [14]. DNMT1 is a maintenance methyltransferase, which is conducive to maintain the methylation pattern. DNMT3A and DNMT3B are the latest methylation enzymes, which participate in the establishment of tissue-specific DNA methylation patterns during development and in response to environmental factors [15]. Secreted frizzledrelated protein (SFRP) is glycoprotein containing a so-called frizzled-like cysteine-rich domain. This domain helps them bind to Wnt ligands or frizzled receptors, regulating Wnt signaling [16]. A previous study has revealed that SFRP2 promotes phosphorylation mediated degradation of β-catenin [17]. Meanwhile, Wnt/β-catenin signaling has been revealed to protect hippocampal neurons from apoptosis [18]. Therefore, this study aims to verify whether METTL3 inhibits autism-like behaviors and hippocampal neuron apoptosis by regulating the MALAT1/SFRP2/Wnt/β-catenin axis.
METHODS
Ethical approval
Animal experimental processes were approved by the Animal Care and Use Committee of Qiqihar Medical university. All experiments were in accordance with the Animal Research: Reporting of In Vivo Experiments guidelines on the Care and Use of Experimental Animals.
Microarray-based gene expression profiling
Differential analysis of the autism-related gene expression dataset GSE38322 retrieved from GEO database was conducted using R “limma” package to screen differentially expressed genes with p<0.05 as the threshold. GSE38322 dataset includes 36 samples, consisting of 18 normal samples and 18 autism samples. Venn diagram was then plotted and m6A-modified genes were screened. Interacting genes of significantly differential m6A-modified genes were predicted through STRING database and a protein-protein interaction (PPI) network was constructed. A confidence score of >0.7 was considered to be statistical significance [19]. Next, Cytoscape was used to plot and calculate the core degree, with genes with degree value >10 considered as hub genes. The differential m6A-modified gene with the highest core degree was selected [20]. Based on the previous reports, its downstream pathways were determined. MEM database was then applied for co-expression analysis to identify the correlation of the m6A-modified gene with downstream genes. The methylases possibly regulated by MALAT1 were predicted by RPISeq. Moreover, the downstream target, SFRP2 was determined based on previous studies and Methprimer database prediction results. STRING database was used to predict the interacting genes of SFRP2, which were then subjected to KEGG pathway enrichment analysis using KOBAS database to identify regulatory pathways.
Establishment of mouse models of autism
Healthy adult ICR male (aged: 8 weeks old; weighing: 40– 70 g) and female mice (aged: 8 weeks old; weighing: 40–60 g) were obtained from Hunan SJA Laboratory Animal Co., Ltd. (Hunan, China). These mice were housed in the SPF laboratory at 25°C and 55% humidity under a 12-h light/dark cycle, with ad libitum access to food and water for acclimatization.
The male and female mice were allowed to mate overnight at a ratio of 2:1. The next morning, the female mice were examined for pregnancy. Then, the pregnant mice were housed in separate cages and randomized into control and valproic acid (VPA)-treated groups. VPA (p4543, Sigma-Aldrich, St. Louis, MO, USA) was dissolved in 0.9% saline at a concentration of 250 mg/mL. Female mice were subjected to a single intraperitoneal injection of 600 mg/kg VPA on E12.5 day after conception, and control female mice were subjected to injection of the same amount of saline at the same time point. The new born mice from the model group were described as autism group; the mice from the control group were described as control group. The offspring were weaned on postnatal day [21]. The following experimentations were carried out only on the male offspring. After 35 days, behavior-related experiments were processed to validate the successful construction of the model.
Animal grouping
Mice after modeling were deeply anesthetized by isoflurane and then injected with lentivirus (Genechem, Shanghai, China) to overexpress or silence METTL3, MALAT1, or SFRP2. Bilateral pores were drilled 1.7 mm posterior and 1.5 mm lateral to the anterior chimney of the mice and the viruses (0.6 μL, 2×106 transducing units/μL) were injected into the hippocampal region at a rate of 0.1 μL per min with the Nanoject II (Drummond Scientific Company, Broomall, PA, USA) system. The injection cannula was slowly retracted 5 min after lentivirus injection. Mice were randomized as follows (8 mice for each group): oe-negative control (NC), oe-MALAT1, oe-NC+sh-NC, oe-METTL3+sh-NC, oe-METTL3+sh-MALAT1, sh-NC, sh-SFRP2 (SFRP2 silencing), sh-SFRP2+dimethyl sulfoxide (DMSO), sh-SFRP2+nitazoxanide (NTZ; Wnt/β-catenin signaling inhibitor), oe-METTL3+DMSO, and oe-METTL3+ NTZ. Wnt/β-catenin signaling inhibitor NTZ was delivered to mice at a dose of 200 mg/kg daily. After 4 weeks, behavioral experiments were performed. Next, the hippocampal tissues of mice were removed and used for biochemical analysis.
Behavioral experiments
The open field test was carried out for assessing the general locomotor activity and anxiety-like behavior utilizing a Plexiglas (100×100×40 cm) [21].
The three-chamber test was a commonly applied method to assay social approach behavior in mice with a 60 cm×40 cm white Plexiglas box. During the test phase, stranger 1 mice of the same sex and same age were randomly placed in metal cages in the left or right box. An unfamiliar object was placed on the metal cage on the other side of the box and the test mice were allowed to freely explore the three experimental compartments for 10 min. Video recordings were used to detect the residence time of the mice in each of the three experimental compartments, and to measure the time the mice spent in establishing social communication with stranger 1. The duration of direct contact between mice and stranger 1 or an empty metal cage was determined. With 3–5 cm around the metal cage defined as the contact range, the duration of mice entering each box was recorded. The head and four claws of mice entering a box was indicative of their existence in the box.
Self-grooming test is performed to analysis repetitive performance of mice in restricted space utilizing a 500-mL glass beaker (7.5 cm diameter×10 cm tall) [21].
TUNEL assay
TUNEL staining kit (In Situ Cell Death Detection Kit; Roche, Basel, Switzerland) was applied for detection of neuron apoptosis in the hippocampal tissues of mice. Briefly, hippocampal tissue sections were incubated with anti-NeuN antibody (1:500, ab177487; Abcam, Cambridge, UK), and then stained with DIPA. For NC, after TUNEL staining, slides were incubated with the TUNEL labeling solution without terminal transferase. The data were expressed as the number of total TUNELand NeuN double-positive per 250 μm length of medial CA1. All measurements were implemented in a blinded manner. There sections were selected from each mouse and then observed under a fluorescence microscope (Olympus Optical Co., Ltd, Tokyo, Japan) in three fields of the CA1 region for cell counting. The number of TUNEL and NeuN double positive cells is the number of cells obtained by counting the field of view in the CA1 region of 250 μm in length [22].
Immunohistochemistry
The hippocampal tissues were fixed in 10% formaldehyde, paraffin-embedded and sliced into 4-μm-thick sections. The tissue sections were heated in a 60°C oven for 1 h, dewaxed, dehydrated and incubated with 3% H2O2 (Sigma-Aldrich) for 30 min. Afterwards, the sections were boiled in 0.01 M sodium citrate for 20 min at 95°C and blocked with normal goat serum (C-005; HaoranBio, Shanghai, China) at 37°C for 10 min. The sections were immunostained with anti-BAX antibody (Rabbit, 1:250, ab32503; Abcam) for 24 h at 4°C. The next day, the sections were incubated with biotin-labelled secondary antibody goat anti-rabbit (1: 500, ab6721; Abcam) for 10 min at room temperature. Afterwards, the sections were treated with horseradish peroxidase (HRP)-labeled streptavidin working solution (0343-10000U; Emmer Biotechnology Co., Ltd., Beijing, China) and developed with DAB (ST033; Guangzhou Weijia Technology Co., Ltd., Guangzhou, China). The sections were then counterstained with hematoxylin (PT001; Shanghai Bogoo Biotechnology Co., Ltd., Shanghai, China) for 1 min, dehydrated, cleared and sealed before observation under an optical microscope. BAX-positive expression is mainly exhibited by yellow-brown staining in the cytoplasm.
Primary mouse hippocampal neuron culture and transduction
The neonatal mice within 24 h were sterilized with 75% medical alcohol, after which the brain of the mice was cut out and the hippocampal tissue was quickly separated, and cut into small pieces. The hippocampal tissue pieces were digested with 0.125% trypsin for 15 min at 37°C, ground and centrifuged. Cells were seeded in 10-mm dishes coated with polylysine (10 mmol/L) at a density of 1×106 cells/mL and maintained in Neural Basal Medium (Gibco, Carlsbad, CA, USA) with 2% B27 (Gibco) and 0.25% Glumax (Gibco). After 3 days, 2.5 μg/mL cytarabine (Sigma-Aldrich) was added to the medium for 24 h to suppress the proliferation of glial cells. Next, 50% of the medium was renewed every three days. Cells were cultured at 37°C with 5% CO2 for 14 days before experimentations [23].
Hippocampal neurons were then transduced with lentivirus (Genechem; multiplicity of infection=5) carrying sh-NC, sh-MALAT1-1, sh-MALAT1-2, sh-MALAT1-3, oe-NC, oe-MALAT1, sh-METTL3-1, sh-METTL3-2, sh-METTL3-3, and oe-METTL3. After 48 h, cells were harvested for subsequent experimentations.
Immunofluorescence
On the 7th day of in vitro culture, primary mouse hippocampal neurons were fixed with 4% paraformaldehyde for 30 min, treated with 0.2% Triton X-100 at ambient temperature for 15 min, blocked with 3% bovine serum albumin (BSA) at 4°C for 30 min, and then incubated with mouse anti-NeuN (ab104224, 1:200; Abcam) in a wet box at 4°C overnight. Next, the cells were incubated with goat anti-mouse secondary antibody (ab150113, 1:200) at ambient temperature away from light for 2 h. 4’,6-diamidino-2-phenylindole (ab104139, 1:100; Abcam) was added for incubation at ambient temperature for 10 min. The results were observed under an inverted fluorescence microscope.
Reverse transcription-quantitative polymerase chain reaction
Total RNA was extracted from mouse hippocampal tissues and cells using a Trizol kit (Thermo Fisher Scientific Inc.). RNA was then reversely transcribed into cDNA employing Reverse Transcription kit (ABI, Applied Biosystems). Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was processed utilizing an ABI7500 quantitative PCR instrument. GAPDH was used as a loading control and the 2-ΔΔCt method was employed to calculate the relative expression of target genes. Primers are provided by Sangon Biotech Co., Ltd. (Shanghai, China) and listed in Supplementary Table 1 (in the online-only Data Supplement).
Western blot
The extracted protein sample was separated using freshlyprepared sodium dodecyl sulphate polyacrylamide gel electrophoresis, electrotransferred onto polyvinylidene fluoride membranes, blocked with 5% BSA for 2 h, and probed with primary antibodies overnight at 4°C. The membranes were then re-probed with HRP-conjugated secondary antibody goat anti-rabbit (ab6721, 1:2,000; Abcam) at room temperature for 1 h. Next, enhanced chemiluminescence reagent (EMD Millipore, Billerica, MA, USA) was used to visualize the results by X-ray film. Protein bands were assayed utilizing Image J software, with β-actin selected for normalization. Primary antibodies used: SFRP2 (12189-1-AP, 1:1,000; Proteintech, Chicago, IL, USA), β-catenin (ab16051, 1:2,000, Abcam), p-β-catenin (ab75777, 1:500; Abcam), BAX (ab32503, 1:1,000; Abcam), Bcl-2 (ab196495, 1:1,000; Abcam), and β-actin rabbit polyclonal antibodies (ab8227, 1:2,000; Abcam).
Methylation specific polymerase chain reaction
Genomic DNA from neurons were extracted with TIANamp Genomic DNA Kit (Tiangen Biotech, Beijing, China), converted by bisulfite, and stored at -80°C. MS-PCR was processed utilizing a methylation-specific kit (Tiangen Biotech) on purified DNA with methylated primers and unmethylated primers (Sangon) described in Supplementary Table 2 (in the online-only Data Supplement). PCR conditions consisted of 10 min at 95°C for initial denaturation, followed by 35 cycles of 95°C (45 s), annealing temperature (methylated [58°C] and unmethylated [57°C] of primers [30 s], and 72°C [45 s]) and a final elongation of 10 min at 72°C. The resultant products were visualized by agarose gel electrophoresis and analyzed by Image J.
RNA binding protein immunoprecipitation assay
The interaction between MALAT1 and indicated DNMTs were validated by RNA binding protein immunoprecipitation (RIP) kit (EMD Millipore). Cells were rinsed by pre-cold phosphate phosphate-buffered saline, lysed by equal volume lysis buffer for 5 min, and centrifuged at 8,000 g for 10 min. The supernatant was incubated with indicated beads pre-conjugated with indicated antibody overnight at 4°C. The samples were harvested in magnetic constellation and then digested by proteinase K and subjected to RNA extraction for following RT-qPCR. Used antibody: DNMT1 (10411-1-AP, 1:50; Proteintech), DNMT3A (20954-1-AP, 1:50; Proteintech), and DNMT3B (MA5-16165, 1:50; Invitrogen Inc., Carlsbad, CA, USA).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assay was implemented employing ChIP Kit (EMD Millipore). Cells were fixed with 1% formaldehyde for 10 min to generate DNA-protein cross-linking and incubated with 0.125 M glycine for 5 min. Then, the cells were lysed and subjected to ultrasonic treatment at 120 w, 2 s on, and 5 s off for 15 cycles to produce appropriately sized fragments. The lysates were incubated antibodies against RNA polymerase II (positive control), IgG (negative control; ab205718, 1:100; Abcam), DNMT1 (10411-1-AP, 1:50; Proteintech), DNMT3A (20954-1-AP, 1:50; Proteintech), and DNMT3B (MA5-16165, 1:50; Invitrogen) at 4°C overnight. Protein agarose/sepharose was used to immunoprecipitate the endogenous DNA-protein complex, which was incubated at 65°C overnight to relieve cross-linking. Phenol/chloroform was then applied for extraction of the obtained DNA fragments. Finally, the DNA fragments were analyzed using qPCR.
Me-RNA binding protein immunoprecipitation assay
Total RNA was isolated from neurons by TRIzol and mRNA was isolated using PolyATtract® mRNA Isolation Systems (AZ5300; Aidlab, Beijing, China). Anti-m6A antibody (Abcam, ab151230) or anti-IgG (ab109475, 1:100; Abcam) was prebound to protein A/G magnetic beads (Pierce) in IP buffer (20-mM Tris pH 7.5, 140-mM NaCl, 1% NP-40, and 2-mM EDTA) for 1 h. mRNA and magnetic beads complex was isolated and purified by IP buffer supplemented with ribonuclease inhibitors and protease inhibitors. RNA was eluted by wash buffer and purified by phenol-chloroform extraction. Primer sequences are described in Supplementary Table 1 (in the online-only Data Supplement). MALAT1 expression was assayed by RT-qPCR.
Photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation
Neurons were incubated with 200 mM of 4-thiopyridine (4SU) (Sigma-Aldrich) for 14 h and crosslinked with 0.4 J/cm2 at 365 nm. The lysates were followed by immunoprecipitation with METTL 3 antibody at 4°C (ab240595, 1:200; Abcam), RNA was precipitated by [g-32-P]-ATP labeling and visualized by radioautocytography. Photoactivatable-ribonucleoside-enhanced-fragments were digested by protease K to remove proteins, and RNA was extracted for RT-qPCR to detect MALAT1 expression levels. The experiment was repeated three times.
Statistical analysis
Data are described as the mean±standard deviation from at least three independent experiments. Statistical comparison was assayed using unpaired t test when only two groups were compared or by Tukey’s test-corrected one-way analysis of variance (ANOVA) when more than two groups were compared. All statistical analyses were implemented with SPSS 20.0 software (IBM Co., Armonk, NY, USA; *p<0.05, **p<0.01, ***p< 0.001, ****p<0.0001).
RESULTS
Bioinformatics analysis predicts that the METTL3/MALAT1/SFRP2/Wnt/β-catenin axis may be involved in the regulation of autism
Differential analysis of the GSE38322 dataset revealed 5,860 differential genes, consisted of 2,748 upregulated and 3,112 downregulated genes. A previous study has suggested that m6A modifications may be related to axonal regeneration in the nervous system [24]. We compared differential genes with m6A-modified genes and found that four m6A-modified genes were significantly differentially expressed in autism (Figure 1A). Next, we predicted five related genes of these four genes by STRING database and constructed a PPI network (Figure 1B). Based on this, we found the highest degree of METTL3 core in the network, which indicated METTL3 might be the key m6A-modified gene in autism. Meanwhile, analysis of the GSE38322 dataset found that METTL3 was significantly downregulated in autism samples (Figure 1C).
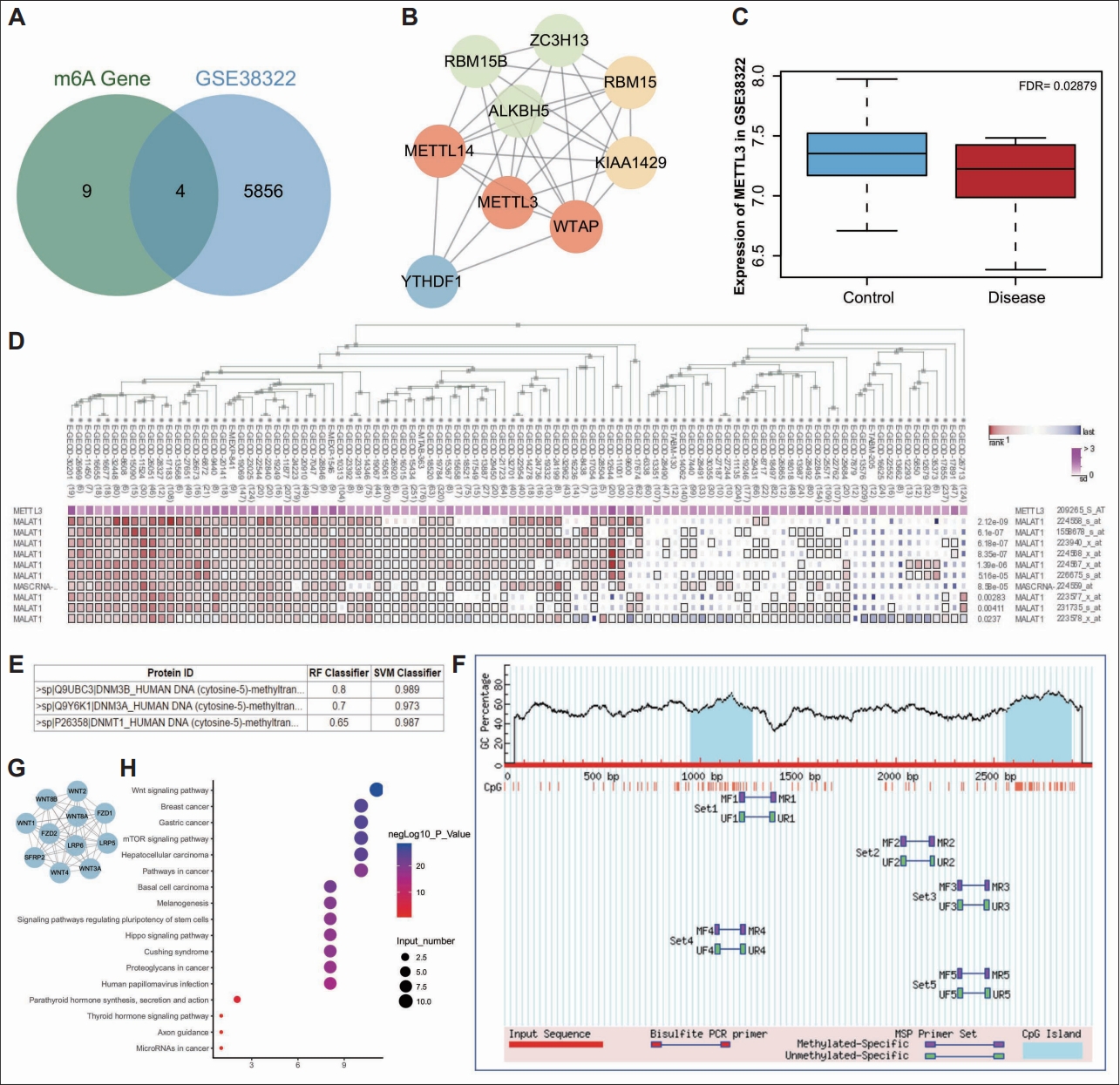
New regulatory pathways in autism predicted by bioinformatics analysis. A: Venn diagram of the significantly differentially expressed genes in autism samples in the GSE38322 dataset and the m6A-modified genes. B: A PPI network of the related genes of RBM15, METTL3 ALKBH5, and YTHDF1 predicted by STRING database. The redder circle reflects higher core degree of the gene and the bluer circle reflects lower core degree of the gene. C: A box plot of METTL3 expression in the GSE38322 dataset. The blue box represents METTL3 expression in normal samples and the red box represents METTL3 expression in autism samples. D: Co-expression correlation between METTL3 expression and MALAT1 expression analyzed by MEM database. E: Interaction between DNMT and MALAT1 predicted by RPISeq database. F: CpG island on SFRP2 promoter predicted by Methprimer database. G: A PPI network of the related genes of SFRP2 predicted by STRING database. H: KEGG enrichment analysis of SFRP2 and its related genes analyzed by KOBRAS database. The ordinate indicates the enriched entry identifiers and the abscissa indicates the number of genes enriched in the entry identifier. The bluer bubble indicates a higher significance while the redder color indicates a lower significance. PPI, protein-protein interaction.
As previously reported, METTL3 can stabilize MALAT1 expression by mediating m6A modification of it [25]. Besides, impaired spatial learning memory was found in METTL3 depleted mice [26]. Moreover, we determined that METTL3 was significantly co-expressed with MALAT1 by co-expression analysis using MEM (Figure 1D). The online website RPISeq predicted that MALAT1 could bind DNA methylation enzymes DNMT1, DNMT3A, and DNMT3B (Figure 1E), and the Methprimer database predicted the presence of large number of CPG islands in the SFRP2 promoter (Figure 1F). What’s more, previous study has documented that SFRP2 can undergo hypermethylation [27]. Thus, we speculated that MALAT1 could recruit DNA methylation enzymes (DNMT3B, DNMT3A, and DNMT1) to regulate the methylation status of SFRP2 in hippocampal neurons of the mouse model of autism. We used STRING database to predict the 10 interacting genes of SFRP2 (Figure 1G), and then the KEGG pathway enrichment of SFRP2 and its interacting genes by KOBAS obtained a total of 17 KEGG pathways, of which the most enriched signaling was Wnt signaling (Figure 1H). Published literature has shown that SFRP2 inhibits the Wnt/β-catenin signaling to promote neuron apoptosis [28], while activation of the Wnt/β-catenin signaling can inhibit neuron damage [29]. SFRP2 may regulate neuron apoptosis in autism via the Wnt/β-catenin signaling. Taken together, we speculated that METTL3 might affect SFRP2 methylation to govern the Wnt/β-catenin signaling by regulating MALAT1 expression via m6A modification, thus influencing neuron apoptosis in autism.
MALAT1 is downregulated in the hippocampal neurons of a mouse model of autism
Then, we moved to determine the role of the aforementioned METTL3/MALAT1/SFRP2/Wnt/β-catenin axis in autism in vivo. We established a mouse model of autism and performed behavioral experiments when mice were 35 days old to validate the success of the model. In the open filed test, the autistic mice exhibited reduced duration and entries in the central area in comparison with control (Figure 2A). Meanwhile, the three chambers social test was carried to determine the social ability, and the results revealed that the autistic mice tended to stay longer time in non-social chamber than in social chamber in contrast to control mice (Figure 2B). Consistently, autistic mice spend more time in repetitive self-grooming than control mice (Figure 2C). Collectively, these data indicated the successful establishment of autism mouse models.

Aberrant downregulation of MALAT1 was found in the hippocampal neurons from mouse models of autism. A: Duration and times of entries in the central area of control and a mouse model of autism determined by open field test. B: Social community capacity of control and a mouse model of autism determined by three chambers test. C: Self-grooming time of control and a mouse model of autism determined by self-grooming test. D: Neuron apoptosis in the CA1 hippocampal tissues of control and a mouse model of autism determined by NeuN staining and TUNEL staining (400×). E: MALAT1 expression in hippocampal tissues of control and a mouse model of autism determined by reverse transcription-quantitative polymerase chain reaction. Data were shown as the mean±standard deviation. The comparison of the two groups of data was conducted using unpaired t test. N=8 mice for each treatment. ***p<0.001; ****p<0.0001.
The hippocampal tissues of mice were collected after behavioral experiments. Neurons were labeled with NeuN and TUNEL staining. The results revealed higher apoptosis rate of hippocampal neurons in the CA1 hippocampal tissues of autistic mice (Figure 2D).
In addition, the results of RT-qPCR suggested that MALAT1 was under-expressed in hippocampal tissues of autistic mice (Figure 2E). Taken together, our data revealed decreased MALAT1 expression in hippocampal tissues of autistic mice.
Ectopic MALAT1 suppresses autism-like behaviors and hippocampal neuron apoptosis in a mouse model of autism
The above findings of aberrant downregulation of MALAT1 in hippocampal tissues of a mouse model of autism allowed us to investigate the role of MALAT1 in autism. MALAT1 was overexpressed in hippocampal tissues of autistic mice and the overexpression efficiency was confirmed by RT-qPCR (Figure 3A).
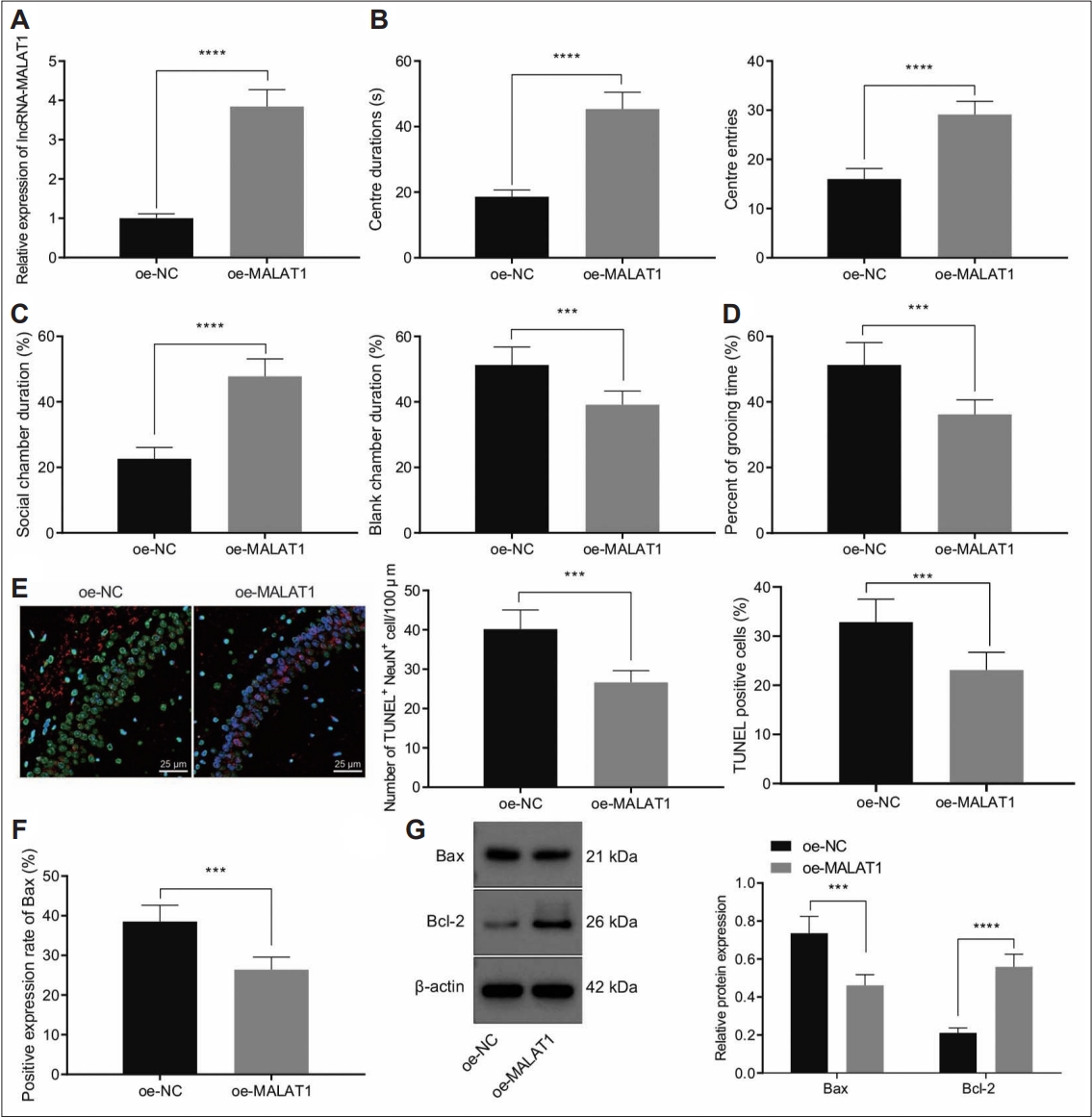
Ectopic MALAT1 expression suppressed autism-like behaviors and hippocampal neuron apoptosis in a mouse model of autism. A: MALAT1 expression in hippocampal tissues of a mouse model of autism treated with oe-MALAT1 determined by reverse transcriptionquantitative polymerase chain reaction. B: Duration and times of entries in central area of a mouse model of autism treated with oe- MALAT1 determined by open field test. C: Social community capacity of a mouse model of autism treated with oe-MALAT1 determined by three chambers test. D: Self-grooming time of a mouse model of autism treated with oe-MALAT1 determined by self-grooming test. E: The neuron apoptosis in the CA1 hippocampal tissues of a mouse model of autism treated with oe-MALAT1 determined by NeuN and TUNEL staining (400×). F: BAX expression in the hippocampal tissues of a mouse model of autism treated with oe-MALAT1 determined by IHC. G: BAX and BCL2 expression in the hippocampal tissues of a mouse model of autism treated with oe-MALAT1 determined by western blot. Data were shown as the mean±standard deviation. The comparison of the two groups of data was conducted using unpaired t test. N=8 mice for each treatment. ***p<0.001; ****p<0.0001. NC, negative control.
Open field test results revealed that MALAT1 overexpression significantly increased the duration and entries in central area of a mouse model of autism (Figure 3B). Moreover, three chambers test data demonstrated autistic mice treated with oe-MALAT1 stayed longer in social chamber than non-social chamber (Figure 3C). Consistently, autistic mice treated with oe-MALAT1 spent less time on self-grooming (Figure 3D). These results indicated that overexpression of MALAT1 alleviated the autism-like behaviors in mice.
NeuN and TUNEL staining results indicated that ectopic expression of MALAT1 reduced neuron apoptosis rates in the CA1 hippocampal tissues of a mouse model of autism (Figure 3E). In addition, MALAT1 overexpression led to decreased BAX positive expression rates in hippocampal tissues of a mouse model of autism (Figure 3F). Meanwhile, western blot results demonstrated that overexpression of MALAT1 increased anti-apoptosis protein BCL2 expression while inhibiting BAX expression (Figure 3G).
Collectively, overexpression of MALAT1 suppressed autism-like behaviors and hippocampal neuron apoptosis in a mouse model of autism.
METTL3 stabilized MALAT1 by promoting m6a modification
A previous report revealed that METTL3 could stabilize MALAT1 by enhancing m6A modification [25]. Hence, we aimed to determine whether METTL3 can promote m6A modification of MALAT1 in autistic mice. RT-qPCR results showed that METTL3 was downregulated in the hippocampal tissues of autistic mice (Figure 4A). Moreover, Me-RIP assay data revealed that m6A level of MALAT1 was reduced in hippocampal tissues of a mouse model of autism (Figure 4B). Immunofluorescence for determination of the expression of neuron marker NeuN showed that the percentage of NeuN positive cells, that is, the neurons, was more than 95% (Figure 4C), which could be used in subsequent experiments.
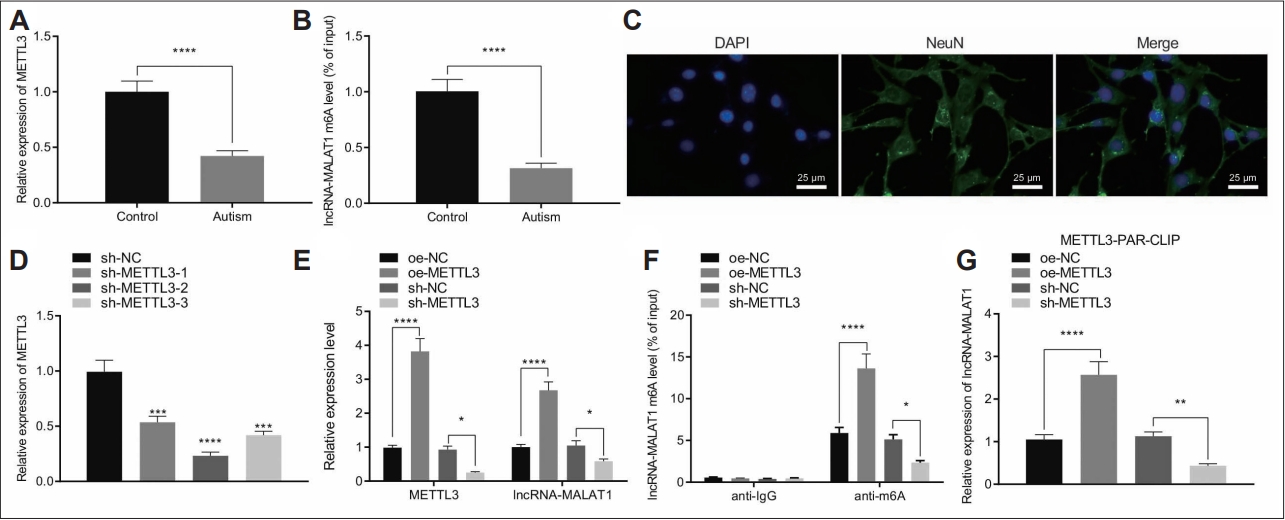
METTL3 promoted the m6A level of MALAT1 to increase the expression of MALAT1. A: METTL3 expression in the hippocampal tissues from control and a mouse model of autism determined by RT-qPCR (N=8). B: m6A levels of MALAT1 in the hippocampal tissues from control and a mouse model of autism determined by Me-RIP (N=8). C: Neuron marker NeuN expression in primary mouse hippocampal neurons determined by Immunofluorescence assay (400×). D: METTL3 silencing efficiency in primary mouse hippocampal neurons determined by determined RT-qPCR. E: METTL3 and MALAT1 in hippocampal neurons treated with oe-METTL3 or sh-METTL3 determined by determined RT-qPCR. F: m6A levels of MALAT1 in hippocampal neurons treated with oe-METTL3 or sh-METTL3 determined by Me- RNA binding protein immunoprecipitation. G: The interaction between METTL3 and MALAT1 in hippocampal neurons treated with oe-METTL3 or sh-METTL3 determined by PAR-CLIP. Data were shown as the mean±standard deviation. Statistical comparison was performed by Tukey’s test-corrected one-way analysis of variance when more than two groups were compared. The cell experiment was repeated 3 times. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. NC, negative control; PAR-CLIP, photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Furthermore, we silenced METTL3 gene in neurons by three independent shRNAs, and RT-qPCR was carried to validate the knockdown efficiency (Figure 4D). Among the three shRNAs, shMETTL3#2 showed superior silencing efficiency and was chosen for following experiments.
We overexpressed and silenced METTL3 in primary mouse hippocampal neurons and found that oe-METTL3 significantly increased MALAT1 expression while sh-METTL3 reduced MALAT1 expression (Figure 4E). In addition, ectopic METTL3 expression elevated the m6A levels of MALAT1, while METTL3 silencing caused opposite results (Figure 4F). Importantly, photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation demonstrated that METTL3 overexpression facilitated its interaction with MALAT1, while precipitated MALAT1 was reduced when METTL3 was depleted (Figure 4G).
Altogether, METTL3 promoted MALAT1 expression by enhancing the m6A level of MALAT1.
METTL3 suppressed autism-like behaviors and hippocampal neuron apoptosis by elevating m6A levels of MALAT1 in a mouse model of autism
Considering the aforementioned results, we then sought to investigate whether METTL3 regulated autism-like behaviours and hippocampal neuron apoptosis by MALAT1. RT-qPCR results confirmed that MALAT1 and MALAT1 expression was significantly upregulated in hippocampal tissues of autistic mice treated with oe-METTL3 while further sh-MALAT1 led to decreased MALAT1 expression without affecting METTL3 expression (Figure 5A).
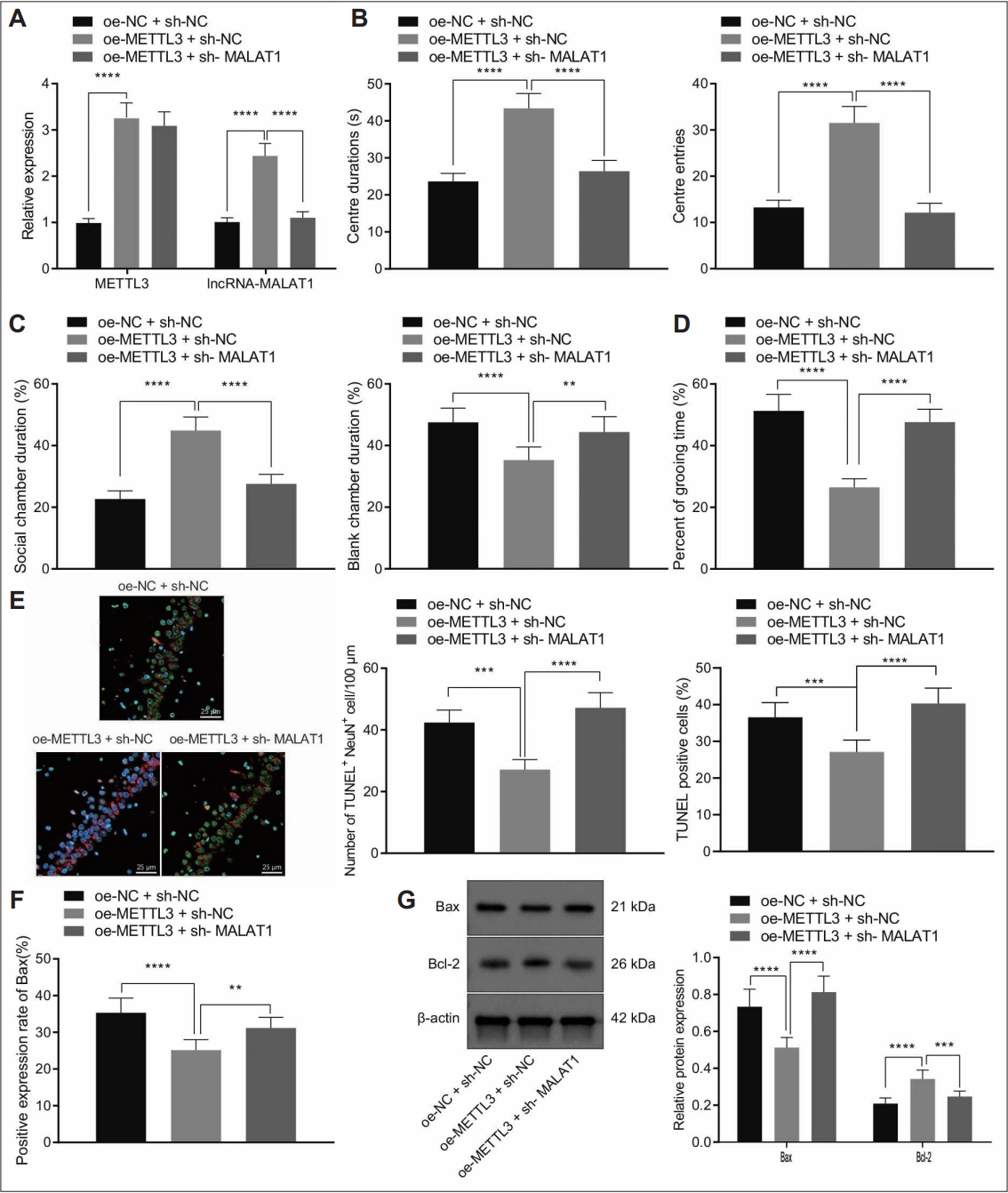
METTL3 promoted m6A modification of MALAT1 to prevent autism-like behaviors and hippocampal neuron apoptosis in a mouse model of autism. A: MALAT1 and METTL3 expression in hippocampal tissues of a mouse model of autism treated with oe-MALAT1 or combined with sh-MALAT1 determined by reverse transcription-quantitative polymerase chain reaction. B: Duration and times of entries in central area of a mouse model of autism treated with oe-MALAT1 or combined with sh-MALAT1 determined by open field test. C: Social community capacity of a mouse model of autism treated with oe-MALAT1 or combined with sh-MALAT1 determined by three chambers test. D: Self-grooming time of a mouse model of autism treated with oe-MALAT1 or combined with sh-MALAT1 determined by self-grooming test. E: The neuron apoptosis in the CA1 hippocampal tissues of a mouse model of autism treated with oe-MALAT1 or combined with sh-MALAT1 determined by NeuN and TUNEL staining (400×). F: BAX expression in hippocampal tissues of a mouse model of autism treated with oe- MALAT1 or combined with sh-MALAT1 determined by immunohistochemistry. G: BAX and BCL2 expression in hippocampal tissues of a mouse model of autism treated with oe-MALAT1 or combined with sh-MALAT1 determined by western blot. Data were shown as the mean±standard deviation. The comparison of the two groups of data was made using the unpaired t test. N=8 mice for each treatment. **p<0.01; ***p<0.001; ****p<0.0001. NC, negative control.
Furthermore, we performed open field test, three chambers test, and self-grooming test to validate mice motor activity and social abilities (Figure 5B-D). The results showed that ectopic METTL3 expression increased the duration and entries in central area in autistic mice. Consistently, autistic mice treated with oe-METTL3 spent more time on social chamber and less time on self-grooming. However, the effect of METTL3 overexpression was reversed by MALAT1 silencing.
Subsequently, NeuN and TUNEL staining results identified that METTL3 overexpression suppressed neuron apoptosis in the hippocampal tissues of autistic mice, while this effect was abolished by MALTA1 depletion (Figure 5E). In addition, METTL3 overexpression dramatically reduced BAX positive expression rates in the hippocampal tissues of a mouse model of autism, while increased BAX positive expression rates were noted due to further MALAT1 silencing (Figure 5F). Similarly, METTL3 overexpression suppressed BAX expression and promoted BCL-2 expression in hippocampal tissues of autistic mice but these results were rescued by depletion of MALAT1 (Figure 5G).
Collectively, our data demonstrated METTL3 reduced autism-like behaviors and hippocampal neuron apoptosis by enhancing m6A level of MALAT1.
MALAT1 promoted methylation of SFRP2 and downregulated its expression
We then investigated the downstream factor of MALAT1 in autism. The results of western blot and RT-qPCR revealed that SFRP2 was upregulated in the hippocampal tissues of a mouse model of autism (Figure 6A and B). In addition, we silenced MALAT1 in hippocampal neurons by three independent shRNAs and RT-qPCR validated the knockdown efficiency (Figure 6C). Among those shRNAs, silencing efficiency of sh-MALAT1#1 was the best and it was chosen for following experiments. Furthermore, overexpression of MALAT1 reduced SFRP2 expression, while its depletion increased SFRP2 expression in neurons (Figure 6D and E).
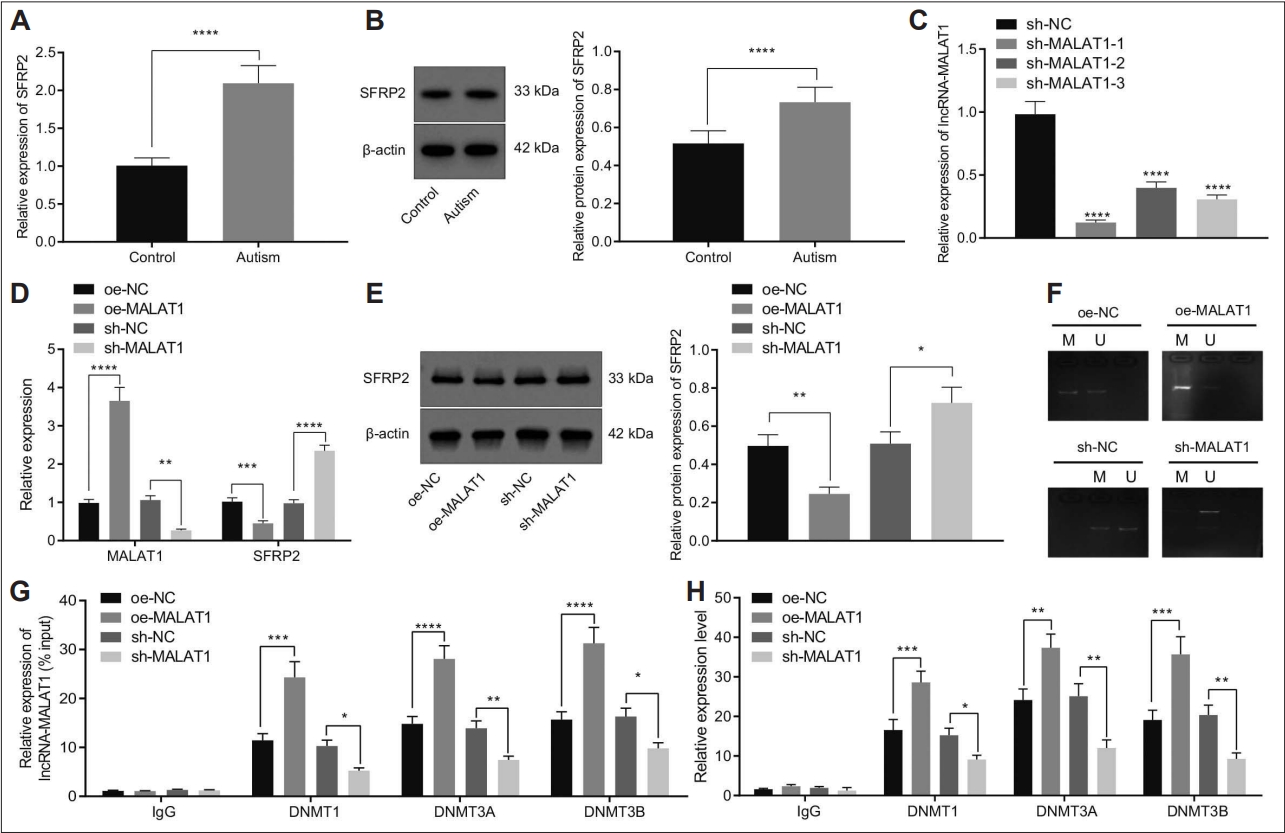
MALAT1 induced methylation of SFRP2 and decreased SFRP2 expression by recruiting DNMT1, DNMT3A, and DNMT3B to the promoter region of SFRP2. A: SFRP2 mRNA expression in the hippocampal tissues of control and a mouse model of autism determined by RT-qPCR (N=8). B: SFRP2 expression in the hippocampal tissues of control and a mouse model of autism determined by western blot (N=8). C: MALAT1 silencing efficiency in hippocampal neurons determined by RT-qPCR. D: SFRP2 and MALAT1 expression in hippocampal neurons treated with oe-MALAT1 or sh-MALAT1 determined by RT-qPCR. E: SFRP2 protein expression in hippocampal neurons treated with oe-MALAT1 or sh-MALAT1 determined by western blot. F: The methylation levels of SFRP2 in hippocampal neurons treated with oe-MALAT1 or sh-MALAT1 determined by MSP assay. G: The interaction between MALAT1 and indicated DNMTs in cell lysates in the presence of oe-MALAT1 or sh-MALAT1 determined by RNA binding protein immunoprecipitation. H: The enrichment of indicated DNMTs on the SFRP2 promoter region in cell lysates in the presence of oe-MALAT1 or sh-MALAT1 determined by chromatin immunoprecipitation. Data were shown as the mean±standard deviation. The comparison of the two groups of data was conducted using unpaired t test. The cell experiment was repeated 3 times. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. NC, negative control; RT-qPCR, reverse transcriptionquantitative polymerase chain reaction.
Moreover, MSP assay results revealed that overexpression of MALAT1 increased the methylation level of SFRP2, while the methylation level was downregulated after MALAT1 inhibition (Figure 6F). The results of RIP assay suggested that overexpression of MALAT1 increased DNMT1, DNMT3A, and DNMT3B-precipetated MALAT1, while it was reduced when MALAT1 silenced (Figure 6G), which indicated MALAT1 interacted with DNMT1, DNMT3A, and DNMT3B.
Furthermore, ChIP assay revealed that overexpression of MALAT1 increased the enrichment of DNMT1, DNMT3A, and DNMT3B on the SFRP2 promoter, while this increase was reversed following MALAT1 silencing (Figure 6H). Collectively, MALAT1 promoted the methylation of SFRP2 and repressed SFRP2 expression by recruiting DNMT1, DNMT3A, and DNMT3B to the promoter region of SFRP2.
SFRP2 inhibited the Wnt/β-catenin signaling to promote autism-like behaviors and hippocampal neuron apoptosis
Previous literature has reported that activation of the Wnt/β-catenin signaling inhibits neuron damage[29] and SFRP2 functions as an antagonist of the Wnt/β-catenin signaling to promote neuron apoptosis [28]. Thus, in the next experiment, we proceeded to analyze whether SFRP2 participates in the apoptosis of hippocampal neurons in autistic mice by affecting the Wnt/β-catenin signaling.
Western blot data showed that β-catenin phosphorylation level was increased in the hippocampal tissues of autistic mice, which led to degradation of β-catenin and a decrease in the protein expression of β-catenin (Figure 7A).

SFRP2 promoted neuron apoptosis by repressing Wnt/β-catenin signaling. A: β-catenin expression in the hippocampal tissues of control and a mouse model of autism determined by reverse transcription-quantitative polymerase chain reaction. B: SFRP2 protein expression in the hippocampal tissues of control and a mouse model of autism determined by western blot. C: β-catenin protein expression and β-catenin phosphorylation levels in the hippocampal tissues of a mouse model of autism treated with sh-SFRP2 or combined with NTZ determined by western blot. D: Duration and times of entries in central area of a mouse model of autism treated with sh-SFRP2 or combined with NTZ determined by open field test. E: Social community capacity of a mouse model of autism treated with sh-SFRP2 or combined with NTZ determined by three chambers test. F: Self-grooming time of a mouse model of autism treated with sh-SFRP2 or combined with NTZ determined by self-grooming test. G: The neuron apoptosis in the CA1 hippocampal tissues of a mouse model of autism treated with sh- SFRP2 or combined with NTZ determined by NeuN and TUNEL staining (400×). H: BAX positive expression in the hippocampal tissues of a mouse model of autism treated with sh-SFRP2 or combined with NTZ determined by immunohistochemistry. I: BAX and BCL2 expression in the hippocampal tissues of a mouse model of autism treated with sh-SFRP2 or combined with NTZ determined by western blot. Data were shown as the mean±standard deviation. The comparison of the two groups of data was conducted using unpaired t test. N=8 mice for each treatment. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. NC, negative control; DMSO, dimethyl sulfoxide; NTZ, nitazoxanide.
Moreover, western blot and RT-qPCR results demonstrated that SFRP2 silencing inhibited β-catenin phosphorylation level and increased β-catenin protein expression, while further NTZ treatment elevated β-catenin phosphorylation level and decreased β-catenin protein expression without affecting SFRP2 mRNA and protein expression (Figure 7B and C).
Furthermore, as shown in Figure 7D-F, SFRP2 silencing increased the duration and entries in central area in autistic mice. Consistently, autistic mice treated with sh-SFRP2 spent more time on social chamber and less time on self-grooming. However, the effect of SFRP2 silencing was reversed by NTZ treatment.
Subsequent NeuN and TUNEL staining results showed that SFRP2 silencing suppressed hippocampal neuron apoptosis in autistic mice, while this effect was abolished by NTZ treatment (Figure 7G). Immunohistochemistry (IHC) data exhibited that SFRP2 silencing reduced BAX positive expression rates, which was reversed by NTZ treatment (Figure 7H).
Similarly, SFRP2 silencing suppressed BAX expression and promoted BCL-2 expression in the hippocampal tissues from autistic mice. In contrast, opposite results were found in response to NTZ treatment (Figure 7I).
Collectively, SFRP2 promoted activation of the Wnt/β-catenin signaling to enhance autism-like behaviors and hippocampal neuron apoptosis in a mouse model of autism.
METTL3/MALAT1/SFRP2 regulated the Wnt/β-catenin signaling to reduce autism-like behaviors and hippocampal neuron apoptosis
Finally, we intended to elucidate the effect of METTL3 regulating the MALAT1/SFRP2/Wnt/β-catenin signaling on the hippocampal neuron apoptosis in vivo. The results of western blot and RT-qPCR revealed that overexpression of METTL3 upregulated MALAT1 and β-catenin expression accompanied with downregulation of SFRP2 expression and β-catenin phosphorylation level. Importantly, ectopic expression of SFRP2 or NTZ treatment in METTL3 overexpressed autistic mice reversed the upregulated β-catenin expression caused by METTL3 overexpression without affecting MALAT1 expression (Figure 8A and B).
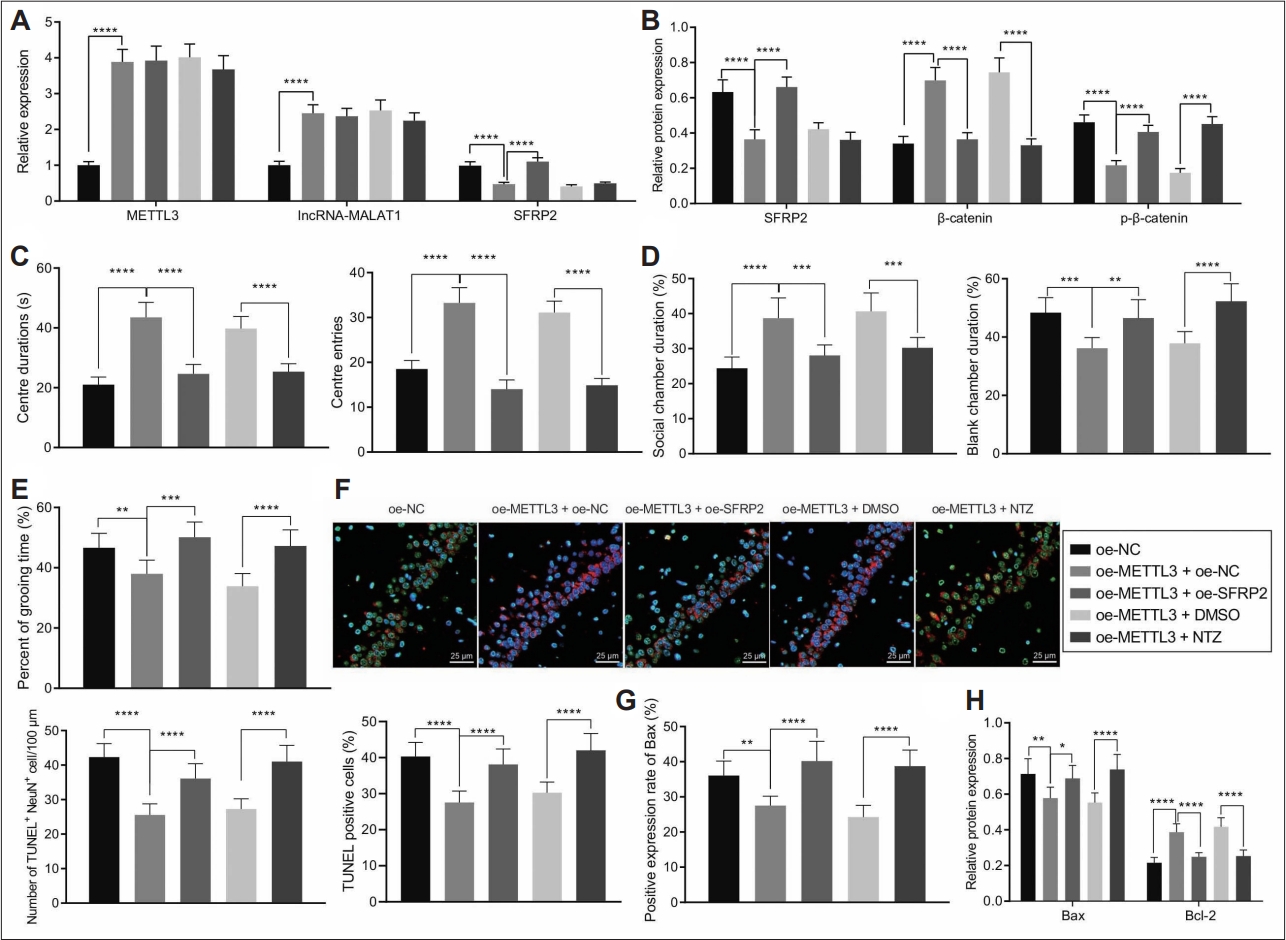
METTL3/MALAT1/SFRP2 regulated the Wnt/β-catenin signaling to regulate autism-like behaviors and hippocampal neuron apoptosis in a mouse model of autism. A: METTL3, MALAT1, and SFRP2 mRNA expression in the hippocampal tissues of a mouse model of autism treated with oe-METTL3, oe-METTL3+oe-SFRP2, or oe-METTL3+NTZ determined by reverse transcription-quantitative polymerase chain reaction. B: METTL3, MALAT1, and SFRP2 protein expression in the hippocampal tissues of a mouse model of autism treated with oe-METTL3, oe-METTL3+oe-SFRP2, or oe-METTL3+NTZ determined by western blot. C: Duration and times of entries in central area of a mouse model of autism treated with oe-METTL3, oe-METTL3+oe-SFRP2, or oe-METTL3+NTZ determined by open field test. D: Social community capacity of a mouse model of autism treated with oe-METTL3, oe-METTL3+oe-SFRP2, or oe-METTL3+NTZ determined by three chambers test. E: Self-grooming time of a mouse model of autism treated with oe-METTL3, oe-METTL3+oe-SFRP2, or oe-METTL3+NTZ determined by self-grooming test. F: The neuron apoptosis in the CA1 hippocampal tissues of a mouse model of autism treated with oe- METTL3, oe-METTL3+oe-SFRP2, or oe-METTL3+NTZ determined by NeuN and TUNEL staining (400×). G: BAX positive expression in hippocampal tissues of a mouse model of autism treated with oe-METTL3, oe-METTL3+oe-SFRP2, or oe-METTL3+NTZ determined by immunohistochemistry. H: BAX and BCL2 expression in hippocampal tissues of a mouse model of autism treated with oe-METTL3, oe- METTL3+oe-SFRP2, or oe-METTL3+NTZ determined by western blot. Data were shown as the mean±standard deviation. Statistical comparison was performed by Tukey’s test-corrected one-way analysis of variance when more than two groups were compared. N=8 mice for each treatment. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. NC, negative control; DMSO, dimethyl sulfoxide; NTZ, nitazoxanide.
Furthermore, the results of open field test, three chambers test, and self-grooming test (Figure 8C-E) found that METTL3 overexpression increased the duration and entries in central area in a mouse model of autism. Consistently, oe-METTL3-treated autistic mice spent more time on social chamber and less time on self-grooming. However, the positive effect of METTL3 overexpression was reversed by ectopic SFRP2 expression or NTZ treatment.
In addition, the results of NeuN and TUNEL staining suggested that METTL3 overexpression suppressed hippocampal neuron apoptosis in a mouse model of autism while this effect was abolished by SFRP2 overexpression and NTZ treatment (Figure 8F).
Consistently, IHC data presented that METTL3 overexpressed reduced BAX positive expression rates, while this reduction was abrogated by SFRP2 overexpression and NTZ treatment (Figure 8G). Similarly, METTL3 overexpression repressed BAX expression and promoted BCL-2 expression in the hippocampal tissues of a mouse model of autism. Importantly, the upregulation of BCL-2 and downregulation of BAX were rescued by SFRP2 overexpression and NTZ treatment (Figure 8H).
Taken together, METTL3/MALAT1/SFRP2 axis prevented autism-like behaviors and apoptosis of hippocampal neuron by regulating the Wnt/β-catenin signaling.
DISCUSSION
Although modification on RNA has been discovered for decades, its roles in the regulation of genes expression are wellstudied until now [6-8]. METTL3 forms complex with METTL14, which responsible for majority m6A modification in cells [30]. Accumulated evidence has demonstrated the vital role of METTL3 in biological processes including metastasis, chemoresistance, DNA repair, and apoptosis [31-33]. Interestingly, a recent study reveals an aberrant downregulation of METTL3 in individuals with autism [10]. However, the molecular mechanism of how METTL3 involved in the regulation of development of autism-like behaviors remains largely unknown. Here, our study firstly analyzed autism-related microarray dataset and found that METTL3 was downregulated in autism samples. Consistent with former studies and microarray data, we further validated significantly downregulated METTL3 expression in hippocampal tissues from a mouse model of autism. What’s more, restoration of METTL3 expression by lentivirus injection in autistic mice alleviated the impaired social communication capacities and motor activity of a mouse model of autism. Notably, our data revealed that ectopic METTL3 expression significantly prevented autism-like behaviors and hippocampal neuron apoptosis.
Moreover, this study revealed that METTL3 could promote MALAT1 expression by facilitating its stability through m6A modification. In agreement with this result, a recent work has identified that METTL3 upregulates MALAT1 expression by promoting its stability via mA modification in the context of IDH-wildtype gliomas [34]. MALAT1 has been determined a significantly differentially expressed gene enriched in the nervous system, immune system and transcription and translationrelated pathways, which appears to be a promising candidate to track clinical improvement following an integrative treatment model in toddlers affected by autism spectrum disorder [35]. MALAT1 has been reported to regulate cell proliferation, metastasis, and chemoresistance [36-38]. Neuron apoptosis is associated with development of autism-like behaviors. Differential penetrance of autism-inducing genetic/epigenetic variants may reflect atypical developmental trajectories associated with Huntingtin functions, including apoptosis [39]. Additionally, neuron apoptosis may also offer an obvious link between autism and glutathione metabolism, which can affect and modulate DNA methylation and epigenetics [40]. MALAT1 has been validated to protect neurons from death [41]. But, its role in the development of autism-like behaviors and hippocampal neuron apoptosis as well as the associated mechanism remains largely unknown and calls for more in-depth investigation.
Accumulating research has uncovered that Wnt/β-catenin signaling is essential for diverse biological processes, including cell cycle transition, cell proliferation, chemoresistance, and apoptosis [42-44]. Importantly, Wnt/β-catenin signaling is tightly involved in the modulation of autism-like behaviors [45]. How the β-catenin is controlled in living cells also has been well studied. For instance, phosphorylation of β-catenin can promote its proteasome dependent degradation [46]. SFRP2, a novel regulator of β-catenin, has been documented to regulate it expression in cardiac fibroblasts [47]. More importantly, our data revealed that SFRP2 was elevated in the hippocampal tissues of autistic mice and negatively correlated with β-catenin protein expression. Furthermore, this study revealed that SFRP2 promoted autism-like behaviors and hippocampal neuron apoptosis by blockage of the Wnt/β-catenin signaling. Notably, our study identified MALAT1 could interact with several DNMTs, DNMT1, DNMT3A and DNMT3B. More importantly, restoration of MALAT1 expression in a mouse model of autism significantly downregulated SFRP2 expression in hippocampal tissues. As far the molecular mechanism of how MALAT1 regulated SFRP2 expression, we revealed that MALAT1 interacted with indicated DNMTs and as a scaffold RNA to recruit the DNMT1, DNMT3A, and DNMT3B to the SFRP2 promoter which led to the hypermethylated CpG island in the SFRP2 promoter and transcription suppression.
Taken together, our data clearly demonstrate METTL3 promotes MALAT1 expression by promoting m6A modification of MALAT1, increases SFRP2 methylation by recruiting DNMT1, DNMT3A, and DNMT3B to the promoter region of SFRP2 and activates the Wnt/β-catenin signaling. Thus, METTL3 represses autism-like symptoms and hippocampal neuron apoptosis (Figure 9). These findings highlight that METTL3 may serve as a novel biomarker for autism. However, further investigation is required based on the clinical specimens.

The mechanism graph of the regulatory network and function of METTL3 in autism. METTL3 stabilizes MALAT1 expression by promoting m6A modification of MALAT1, increases SFRP2 methylation and activates the Wnt/β-catenin signaling. By this mechanism, METTL3 suppresses autism-like symptoms and hippocampal neuron apoptosis.
Supplementary Materials
The online-only Data Supplement is available with this article at https://doi.org/10.30773/pi.2021.0370.
Primer sequences used for reverse transcription-quantitative polymerase chain reaction
Primer sequences used for MS-PCR
Notes
Availability of Data and Material
The data underlying this article will be shared on reasonable request to the corresponding author.
Conflicts of Interest
The authors have no potential conflicts of interest to disclose.
Author Contributions
Conceptualization: Yue Ming, Zhihui Deng. Data curation: Xianhua Tian, Yuerong Jia. Formal analysis: Xianhua Tian, Yuerong Jia. Funding acquisition: Yue Ming, Shuhua Cheng. Investigation: Yue Ming, Zhihui Deng. Methodology: Yue Ming, Zhihui Deng. Project administration: Shuhua Cheng. Resources: Yue Ming. Software: Meng Ning. Supervision: Shuhua Cheng. Validation: Xianhua Tian. Visualization: Shuhua Cheng. Writing—original draft: Meng Ning, Xianhua Tian, Yuerong Jia, Shuhua Cheng. Writing—review & editing: Yue Ming, Zhihui Deng.
Funding Statement
This study was supported by General Program of Basic Scientific Research of Heilongjiang Provincial Department of Education in 2018 (No. 135309433) and Key Topics in 2021 of the 14th Five-Year Plan of Education & Science in Heilongjiang Province (No. GJB1421346).
