Profiling of Proteins Regulated by Venlafaxine during Neural Differentiation of Human Cells
Article information
Abstract
Objective
Antidepressants are known to positively influence several factors in patients with depressive disorders, resulting in increased neurogenesis and subsequent relief of depressive disorders. To study the effects of venlafaxine during neural differentiation at the cellular level, we looked at its effect on protein expression and regulation mechanisms during neural differentiation.
Methods
After exposing NCCIT cell-derived EBs to venlafaxine during differentiation (1 day and 7 days), changes in protein expression were analyzed by 2-DE and MALDI-TOF MS analysis. Gene levels of proteins regulated by venlafaxine were analyzed by real-time RT-PCR.
Results
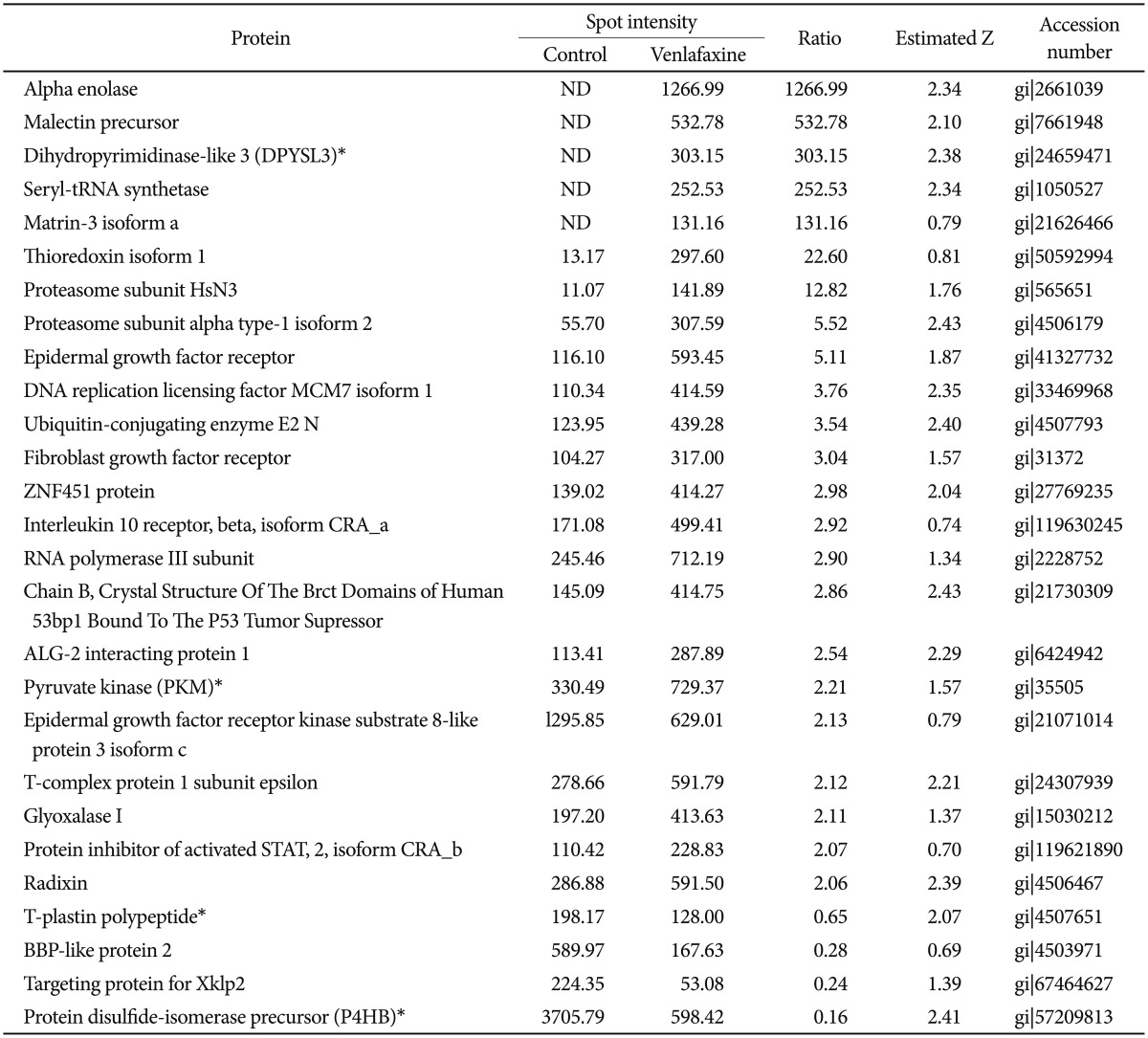
Treatment with venlafaxine decreased expression of prolyl 4-hydroxylase (P4HB), ubiquitin-conjugating enzyme E2K (HIP2) and plastin 3 (T-plastin), and up-regulated expression of growth factor beta-3 (TGF-β3), dihydropyrimidinase-like 3 (DPYSL3), and pyruvate kinase (PKM) after differentiation for 1 and 7 days. In cells exposed to venlafaxine, the mRNA expression patterns of HIP2 and PKM, which function as negative and positive regulators of differentiation and neuronal survival, respectively, were consistent with the observed changes in protein expression.
Conclusion
Our findings may contribute to improve understanding of molecular mechanism of venlafaxine.
INTRODUCTION
Treatment with antidepressants promotes a rapid increase in levels of monoamines in the synaptic cleft, whereas the clinical effects of antidepressants are observed only after several weeks to months.1,2 Previous studies have shown that chronic treatment of antidepressants such as tianeptine and fluoxetine increases expression of brain-derived neurotropic factor (BDNF), mitogen-activated protein kinase, Bcl-xL, and NCAM140, as well as increases cell survival, suggesting that antidepressants affect the cellular and molecular mechanisms in the brain in addition to increasing levels of monoamines.3,4,5,6,7
The mechanisms of antidepressants associated with stress and depression have been persistently studied both in vitro and in vivo. Stress and/or depression suppress neurogenesis in the subgranular zone (SGZ) of the adult hippocampus.8,9 It has been reported that depressive disorders induce atrophy and decrease the volume of the hippocampal region of the brain.10,11 On the other hand, chronic treatment with antidepressants such as fluoxetine results not only in recovery of stress-induced decreases in neurogenesis, but also improves cell proliferation in the SGZ of the adult hippocampus, suggesting the involvement of complex processes such as neural plasticity.9,12,13 Several previous studies have utilized proteomic analysis to evaluate the mechanisms of action of antidepressants.14,15,16 Khawaja et al.14 found that chronic treatment of rats with antidepressants (14 days) including fluoxetine and venlafaxine, induced changes in the expression of proteins in the hippocampus that are involved with cellular proliferation, differentiation, apoptosis, and survival mechanisms. Likewise, Cecconi et al.15 identified differently expressed proteins in rat cortical neurons after fluoxetine treatment for 3 days, which were associated with neuroprotection, serotonin biosynthesis, and axonal transport. Similarly, treatment of mouse embryonic stem (ES) cell-derived neural cells with toparoxetine for 14 days strongly upregulates the expression of proteins related to the synthesis of neurotransmitters.16 Together, these results suggest that antidepressants positively influence neurons both before and after ES cell differentiation to relieve depressive disorders through various molecular and cellular mechanisms.
In particular, venlafaxine is one of serotonin-norepinephrine reuptake inhibitors (SNRI), which increase the levels of both serotonin and norepinephrine in the synaptic cleft by blocking transport of both monoamines. Several studies suggest that venlafaxine is more effective than serotonin selective reuptake inhibitors (SSRIs) such as fluoxetine.17,18 However, there is insufficient evidence to indicate a molecular mechanism of antidepressants associated with various network pathways at the transcriptomic and proteomic levels during neural differentiation.
Human embryonic carcinoma (EC) cells are pluripotent stem cells derived from malignant teratocarcinoma and are considered to be in same category as human ESCs.19,20,21 Importantly, EC cells are easier to handle than ES cells with respect to studying the molecular and cellular mechanisms of cell differentiation during embryonic development. In addition, one of human EC cell line, NCCIT cells are known to induce multilineage differentiation upon exposure to retinoic acid (RA) in vitro.22,23 Gasimli et al.22 observed distinct morphological changes, showing branched elongated process of neuronal morphology after 20 days of RA treatment in NCCIT cells. As a result of aggregates, ES cells or EC cells can spontaneously form embryonic bodies (EBs) in vitro.24 Tonge et al.25 observed that neural gene expression was increased in RA-treated EBs of human ES cells compared to monolayer of cells.
To study the effects of venlafaxine during neural differentiation at the cellular level, we induced differentiation of NCCIT cells by exposing them to RA in the presence or absence of venlafaxine. We identified proteins that were regulated by venlafaxine during neural differentiation using proteomic analysis and examined the mechanisms by which the identified proteins were regulated during differentiation. The proteins that were identified as being regulated by venlafaxine in this study may contribute to a better understanding of the molecular mechanism by which venlafaxine treatment relieves depressive disorders.
METHODS
NCCIT cell culture and embryonic body formation
The NCCIT cell line (CRL-2073) was obtained from the ATCC (Manassa, VA, USA). Cells were cultured in RPMI-1640 medium (Invitrogen, CA, USA) containing 10% fetal bovine serum (Invitrogen), 100 U/mL penicillin, and 10 µg/mL streptomycin (Invitrogen) at 37℃ in a humidified atmosphere with 5% CO2. To induce the formation of EBs, NCCIT cells were transferred to 90-mm bacterial culture dishes for non-adherent culture condition for 7 days.26 The culture medium was replaced every 2 days with fresh medium.
Venlafaxine treatment during neural differentiation of EBs
To evaluate the effects of venlafaxine during neural differentiation, EBs derived from NCCIT cells were seeded in 100-mm tissue culture dishes and incubated with differentiation medium containing 10 µM RA in the absence or presence of 10 µM venlafaxine (Wyeth Korea, Seoul, Korea) (Figure 1A). The 10 µM concentration of venlafaxine was decided as was done in previous studies that the concentration did not induce apoptosis.27,28 Cultures were fed with fresh differentiation medium every 2 to 3 days up to 7 days.
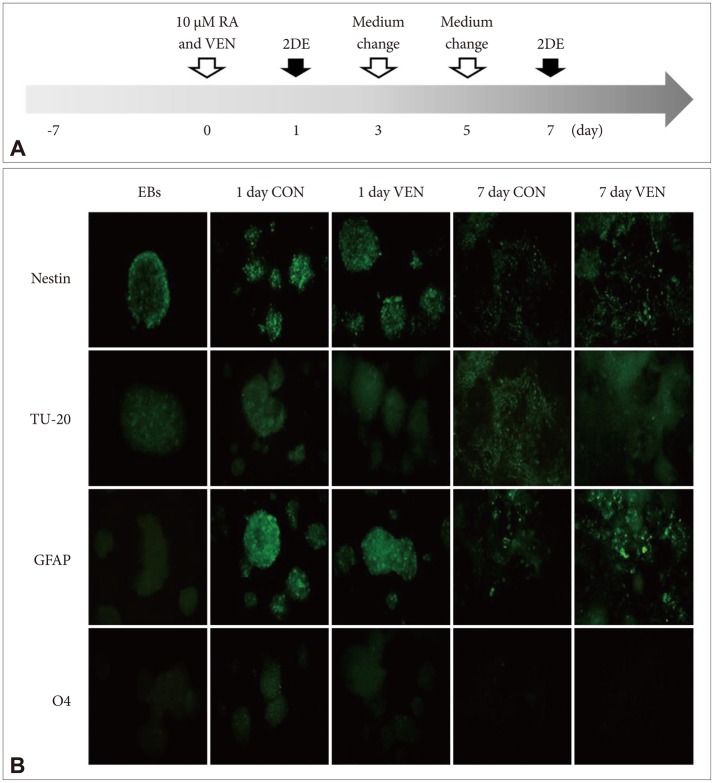
Induction of differentiation of embryonic bodies derived from NCCIT cells. A: Experimental scheme. Human embryonic carcinoma (NCCIT) cells were induced to form embryonic bodies (EBs) for 7 days. EBs were treated with 10 µM retinoic acid (RA) in the presence or absence of 10 µM venlafaxine (VEN) for 7 days. Cells on differentiation day 1 and 7 were collected for two-dimensional gel electrophoresis (2DE) assay. B: Measurement of the differentiation potential of EBs. After exposing EBs to 10 µM RA in the presence or absence of 10 µM VEN for 7 days, immunocytochemistry was used to measure the Nestin (neural stem cell marker), TU20 (neuron marker), GFAP (astrocyte marker), and O4 (oli-godendrocyte marker). Cells exposed to RA in the absence of VEN served as controls (CON).
Immunocytochemistry
EBs derived from NCCIT cells were plated onto cover slips pre-coated with poly-L-lysine (Sigma-Aldrich, MO, USA) in RPMI 1640 medium containing 10% fetal bovine serum and 1% penicillin/streptomycin. After differentiation (1 day or 7 days), the cells were fixed with 4% paraformaldehyde and permeabilized with 100% ice-cold methanol, then washed with PBS containing 0.04% sodium azide and incubated in PBS containing 5% horse serum (Sigma-Aldrich) for 1 h. The cells were incubated with primary antibodies against beta-tubulin III (Tu-20, 1:200) (Millipore, MA, USA), glial fibrillary acidic protein (GFAP, 1:500) (Dakocytomation, Glostrup, Denmark), O4 (1:200) (Chemicon, CA, USA), and Nestin (1:250) (Chemicon) for 1 h at room temperature. The cells were then washed with PBS and incubated with fluorescein-labeled anti-rabbit IgG (H+L) (Jackson ImmunoResearch, PA, USA) and fluorescein-labeled anti-mouse IgG (H+L) (Jackson ImmunoResearch). The stained cells were visualized with a mounting solution containing 4'-6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA, USA) and observed using an ECLIPSE 80i fluorescence microscope (Nikon, Tokyo, Japan).
Two-dimensional gel electrophoresis
For 2D gel electrophoresis, immobilized pH gradient (IPG) dry strips were equilibrated for 12-16 h with reswelling solution containing 7 M urea, 2% 3-[(3-cholamidopropy) dime-thyammonio]-1-propanesulfonate (CHAPS), 1% dithiothreitol (DTT), and 1% pharmalyte. Next, 200 µg of the samples were loaded onto the strip; protein concentrations were determined by Bradford assay (Sigma-Aldrich). Isoelectric focusing (IEF) was carried out at 20℃ using a Multiphore II system (Amersham Biosciences, NJ, USA) and EPS 3500 XL power supply (Amersham Biosciences) according to the manufacturer's instructions. Prior to the second dimension, the focused IPG strips were reduced (1% DTT) and alkalized (2.5% iodoacetamide) in equilibration buffer (50 mM Tris-Cl, pH 6.8 containing 6 M urea, 2% SDS and 30% glycerol). SDS-PAGE was then performed with a Hoefer DALT 2D system (Amersham Biosciences) at 20℃. Two-dimensional gel electrophoresis (2-DE) gels were stained with Coomassie Brilliant Blue dye. Analysis of imaged spots was performed using PD-Quest software (version 7.0, BioRad). To determine the relative levels of protein in each individual spot, the spot volume of each protein was compared to that of a normalized protein volume.
In-gel digestion
Differentially expressed protein spots were enzymatically digested in gel using modified porcine trypsin (Promega, WI, USA) as previously described.29 Briefly, SDS and salts were removed by washing gel pieces with 50% acetonitrile followed by drying to remove the solvent. The washed gels were then rehydrated with trypsin (8-10 ng/µL) and incubated for 8-10 h at 37℃. The proteolytic reaction was terminated by the addition of 20 µL of 0.1% trifluoroacetic acid. Tryptic peptides were recovered by extraction in 50% aqueous acetonitrile, and aqueous phase extractions of replicate gel pieces were combined. An aliquot of the solution was mixed with an equal volume of saturated solution of α-cyano-4-hydroxycinnamic acid in 50% aqueous acetonitrile, and 1 µL of the mixture was spotted on the target plate.
MALDI-TOF MS analysis and database search
Peptide analysis was performed using an Ultraflex MALDI-TOF MS (Bruker Daltonics, Bremen, Germany). Briefly, peptides were evaporated with an N2 laser at 337 nm using a delayed extraction approach and accelerated with a 20 kV injection pulse for TOF analysis. Each spectrum was the cumulative average of 300 laser pulses. Proteins were identified by peptide mass fingerprinting using the ProFound search program developed by Rockefeller University (http://prowl.rockefeller.edu/prowl-cgi/profound.exe). Spectra were calibrated using trypsin autodigestion ion peak signals (m/z 842.510 and 2211.1046) as internal standards.
Pathway network analysis
Integrated pathway enrichment analysis was performed using knowledge-based canonical pathways and the endogenous metabolic pathways of MetaCore pathway analysis software (GeneGo Inc, MI, USA). Briefly, differentially expressed proteins were mapped to biological networks using a manually curated proprietary database (GeneGo) and MetaCore pathway analysis software; gene symbols of differentially expressed proteins were uploaded into the GeneGo database. For enrichment analysis, the gene IDs of the uploaded files were matched with the gene IDs of functional ontologies in MetaCore, which included canonical pathway maps (GeneGo maps), GeneGo cellular processes, Gene Ontology (GO) cellular process, and disease categories. The canonical pathway maps comprised a set of approximately 500 signaling and metabolic maps covering human biology. For network analysis, we utilized the analyze networks, transcription regulation, and direct interactions algorithms. The analyze networks algorithm, a variant of the shortest paths algorithm, uses relative enrichment and relative saturation to generate sub-networks that are prioritized by the number of associated canonical pathways.
Real-time RT-PCR of genes regulated during neural differentiation of EBs exposed to venlafaxine
Total RNA from cells differentiated in the absence or presence of 10 µM venlafaxine for 1 or 7 days was extracted using TRIzol® Reagent (Invitrogen) according to the manufacturer's instructions. Extracted total RNA was reverse transcribed into cDNA using PrimeScript™ Reverse Transcriptase (Takara, Shiga, Japan). Real-time PCR was performed using the 7500 real-time PCR system (Applied Biosystems, CA, USA) with 2X SYBR Green PCR Master Mix (Takara) as previously described.30,31 Reaction mixtures contained 2X SYBR Green PCR Master Mix, 8 pmol each of forward and reverse primer, and 0.5 µL cDNA. During each cycle, the accumulation of PCR products was detected by measuring the increase in fluorescence of a double stranded DNA-binding SYBR Green reporter dye. β-actin was used as an internal control. The sequences of primers are listed in Table 1.
Statistical analysis
All of the data are expressed as the mean±standard error of the mean (SEM) and were analyzed using OriginPro 8 (Origin-Lab Corporation, MA, USA). Statistical analyses were performed using SPSS 17.0 (SPSS Inc., IL, USA). p-values<0.05 were considered significant.
RESULTS
Differentiation potential of EBs after exposure to venlafaxine
To identify the effects of venlafaxine on differentiation of EBs derived from NCCIT cells, we induced differentiation of EBs in the presence or absence of venlafaxine for 1 or 7 days and measured differentiation potential by immunocytochemistry. In EBs derived from NCCIT cells, all of the tested markers related to central nervous system development were expressed after 1 day of differentiation (Figure 1B). The levels of expression of GFAP and Nestin, which are astrocyte and neural stem cell markers, respectively, were decreased for 7 days in both venlafaxine-treated and control cells. On the other hand, the signal intensity of the neuron marker TU-20 in venlafaxine-treated cells was not significantly different compared with control cells. As 7 day-differentiation of EBs derived from NCCIT cells into neurons were at an early stage, the signal intensity of the neuron marker was weak in both venlafaxine-treated and control cells. Thus, at the early stage of neuronal differentiation evaluated in our study, it was difficult to detect the effect of venlafaxine on early stage neuronal differentiation by immunocytochemistry.
Regulation of proteins by venlafaxine during neural differentiation of NCCIT cells
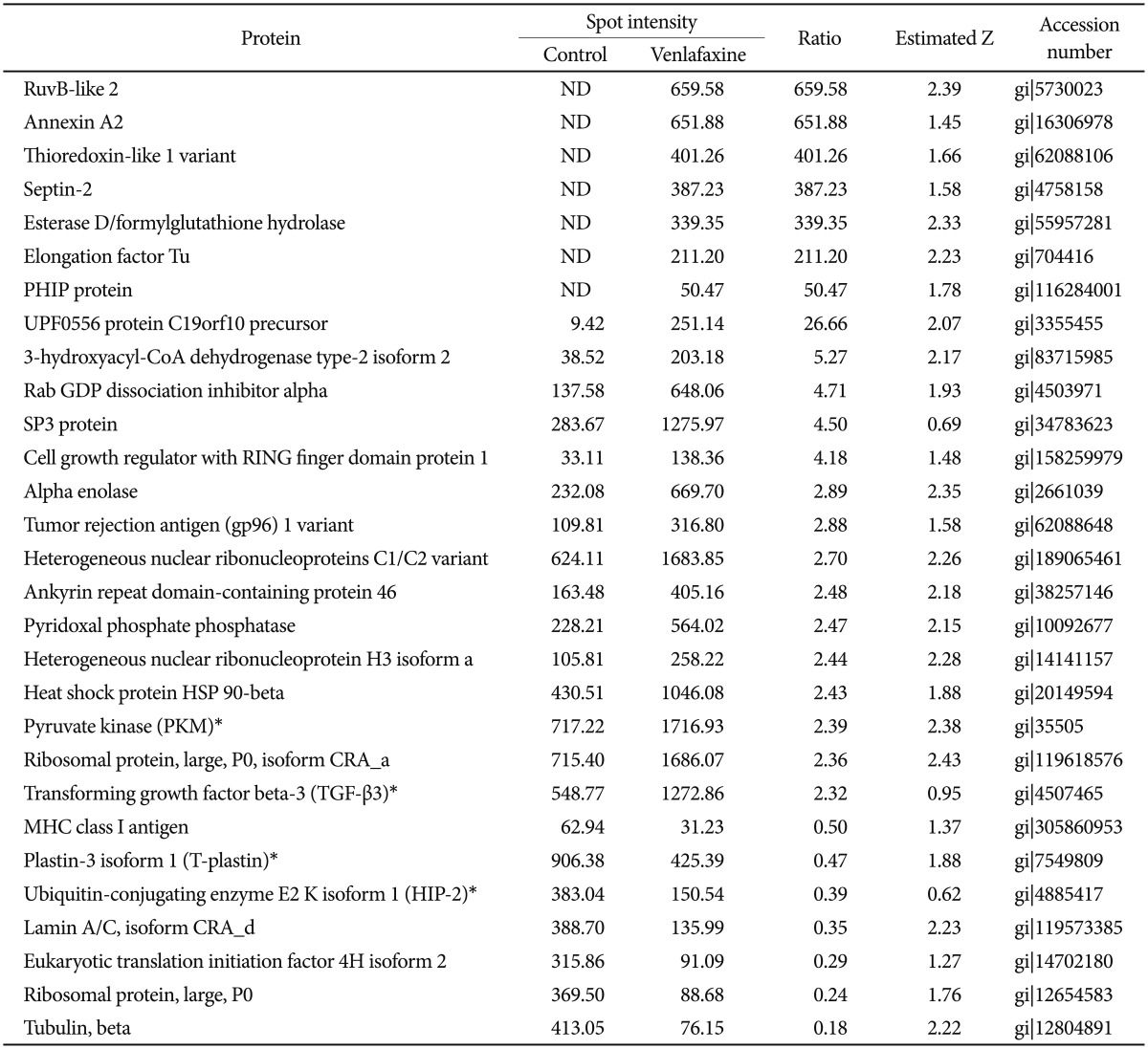
To analyze the proteins regulated by venlafaxine during neural differentiation of NCCIT cells, we performed a 2DE assay after exposing EBs to 10 µM venlafaxine for 1 or 7 days. Of the spots obtained from the 2DE gel, those that were differentially regulated by venlafaxine were analyzed by MALDI-TOF MS. Among the proteins that were up-regulated in cells treated with venlafaxine after differentiation for 1 day, we identified ruvB-like 2 (RUVBL2), elongation factor Tu (TUFM), heat shock protein HSP 90-beta (HSP90AB1), pyruvate kinase (PKM), transforming growth factor beta-3 (TGF-β3), and alpha enolase (ENO1) (Table 2). Likewise, expression of PKM, dihydropyrimidinase-like 3 (DPYSL3), DNA replication licensing factor (MCM7), epidermal growth factor receptor (EGFR), and ENO1 were up-regulated by venlafaxine after differentiation for 7 days (Table 3). TUFM, PKM, TGF-β3, ENO1, DPYSL3, MCM7, and EGFR are associated with cellular growth, differentiation, and proliferation, and thus we considered the possibility that venlafaxine may positively influence neural differentiation. Conversely, ubiquitin-conjugating enzyme E2 K isoform 1 (HIP-2), lamin A/C, eukaryotic translation initiation factor 4H (EIF4H), ribosomal protein, large, P0 (RPLP0), and tubulin beta (TUBB) protein were down-regulated by venlafaxine after differentiation for 1 day (Table 2), while BP-like protein 2 (TM2D3), plastin3 (T-Plastin), and protein disulfide-isomerase precursor (P4HB) were down-regulated by venlafaxine after differentiation for 7 days (Table 3).
The summarized list of up- or down-regulated proteins in the differentiated cells in the presence of venlafaxine for 1 day as compared to the control
Identification of potential networks of proteins regulated by venlafaxine
To explore the GeneGo Process Networks for proteins regulated by venlafaxine during neural differentiation, we used MetaCore database and MetaCore pathway analysis software (GeneGo Inc.). The MetaCore database is well suited for analyzing manually curated interactions and contains >90% of human proteins with known functions.26 We identified top scoring networks that were common to both venlafaxine-treated and control cells during neural differentiation (Figure 2). The highest scoring networks that were regulated by venlafaxine on differentiation for 1 day included TGF-β3, PKM, and HIP-2, which are involved in negative regulation of biological process (63.3%) and regulation of apoptosis (44.7%) (Figure 2A). Likewise, the highest scoring networks that were regulated by venlafaxine on differentiation for 7 days included damage-specific DNA binding protein 1 (DDB1), PKM, and P4HB, which are involved in response to chemical stimulus (76.0%) and regulation of biological quality (60.0%) (Figure 2B).
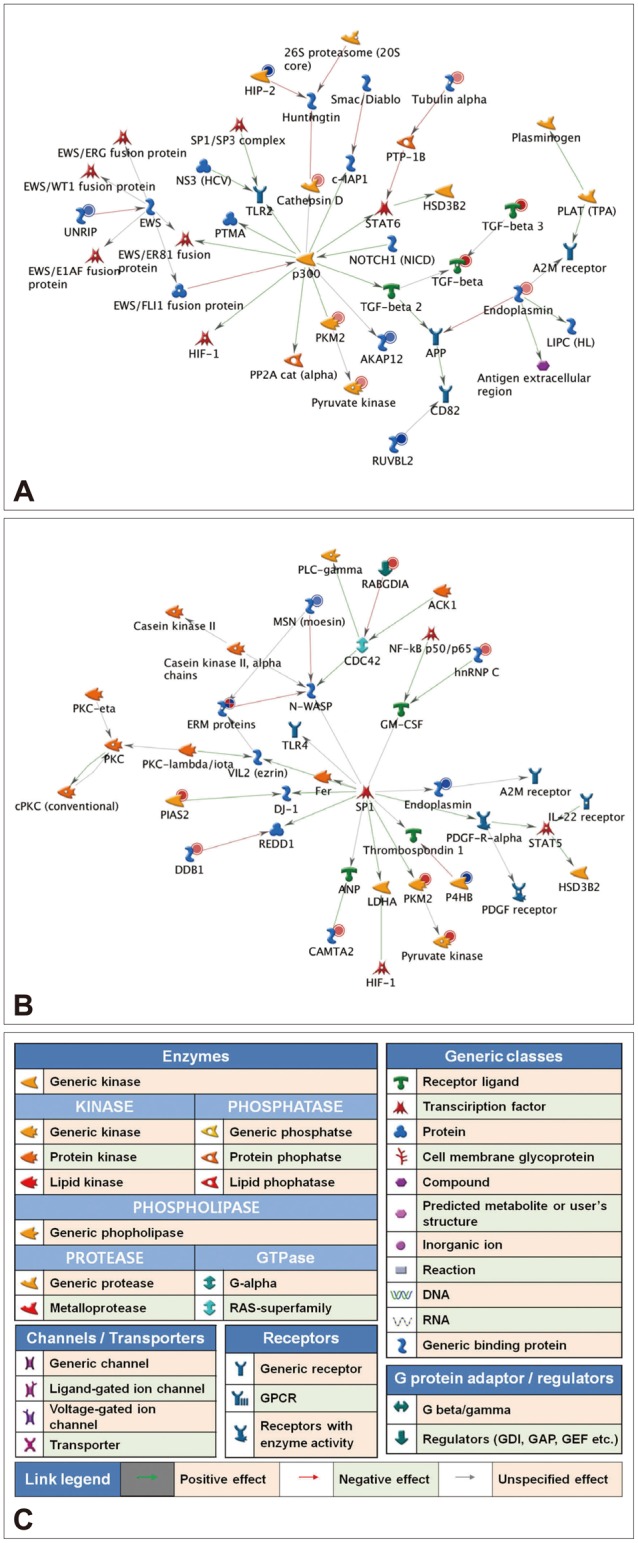
Regulation of protein expression networks by venlafaxine during neural differentiation of NCCIT cells. EBs were treated with 10 µM retinoic acid (RA) in the presence or absence of 10 µM venlafaxine (VEN) for either 1 or 7 days, and putative targets that were differentially regulated by VEN were analyzed using the analyze networks algorithm implemented in MetaCore. A: Network of proteins regulated by VEN after differentiation for 1 day. B: Network of proteins regulated by VEN after differentiation for 7 days. C: Network legend, red and blue circles indicate up- and down-regulated proteins, respectively.
Validation of mRNA levels of protein regulated by venlafaxine
Although post-translational modification and protein turn over can affect the levels of protein expression, determining that an identified protein target is regulated at the mRNA level provides corresponding evidence of the reliability of the relative quantitative protein expression results. Thus, we measured transcript levels of selected proteins using real-time RT-PCR. Similar to the proteomic expression pattern, the expression levels of HIP2 and T-plastin mRNA were decreased while levels of PKM and TGF-β3 mRNA were increased in venlafaxine-treated cells compared to control cells after differentiation for 1 day (Figure 3A and B). After differentiation for 7 days, the levels of expression of P4HB and T-plastin mRNA were significantly decreased, while the expression of PKM and DPYSL3 mRNAs were increased in venlafaxine-treated cells compared to control cells. Together, these results corresponded with the observed patterns of protein expression (Figure 3C and D). On the other hand, although expression of DPYSL3 and HIP2 proteins could not be detected after 1 and 7 days of differentiation, respectively, changes in their expression at the mRNA level were confirmed. Based on these results, it appeared that, beginning almost immediately after initiation of differentiation and up to 7 days later, treatment with venlafaxine increased consistently levels of DPYSL3 and levels of HIP2 gradually decreased. Together, these data served to validate several proteins obtained from proteomic profiling, as well as provided information regarding estimated expression levels of proteins that were identified at only a single time point by proteomic analysis.
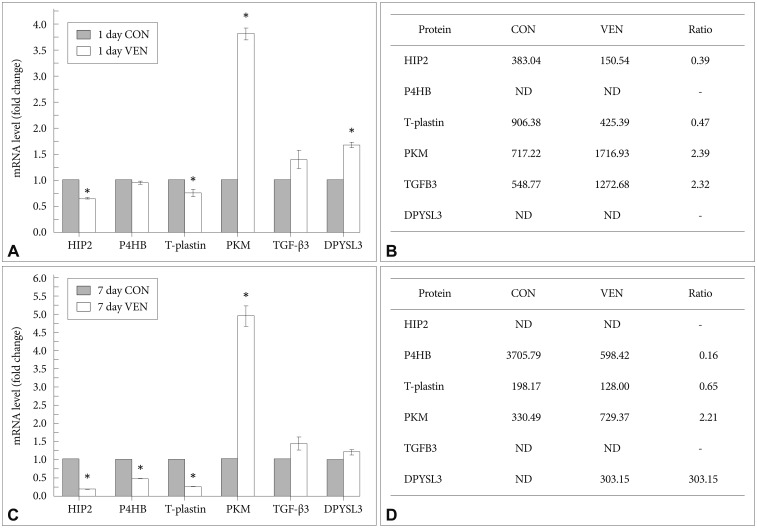
mRNA expression patterns of proteins regulated by venlafaxine during neural differentiation of NCCIT cells. EBs were treated with 10 µM retinoic acid (RA) in the presence or absence of 10 µM venlafaxine (VEN) for 1 or 7 days, and mRNA expression levels of targets differentially regulated by VEN were analyzed by real-time RT-PCR. A: Levels of mRNAs regulated by VEN after differentiation for 1 day. B: Spot intensity of proteins regulated by VEN after differentiation for 1 day. C: Levels of mRNAs regulated by VEN after differentiation for 7 days. D: Spot intensity of proteins regulated by VEN after differentiation for 7 days. Each bar in A and C represents the mean±SEM (N=3). *significantly different from CON by the two-sample t-test (p<0.05). ND: not detected by 2DE assay.
DISCUSSION
Neurogenesis progresses through the interactions of various factors associated with survival, differentiation, metabolism, and proliferation. Antidepressants are known to positively affect these factors in patients with depressive disorders, resulting in increased neurogenesis and subsequent relief of depressive disorders.9,32,33 Therefore, it is meaningful to identify proteins regulated by antidepressants under in vitro conditions, as this information can provide clues about the complex network of interactions that take place between antidepressants and cells in the brain. In the present study, we used proteomic analysis to identify proteins that were differentially regulated by venlafaxine during neural differentiation of NCCIT cells.
The results of our study showed that after 1 and 7 days of neural differentiation of EBs, venlafaxine did not significantly alter expression of the neuronal marker TU-20 at the cellular level. These results were consistent with previous studies showing that that NCCIT cells differentiate into neurons after 3-4 weeks of continuous exposure to RA in vitro22 and administration of rolipram, an antidepressant, to mice for 14 days promotes maturation of new neurons,34 because the maximum treatment with venlafaxine in our study was 7 days.
In the present study, TGF-β3 and DPYSL3 were up-regulated after 1 and 7 days of differentiation and treatment with venlafaxine. The TGF-β subfamily comprises TGF-β1, 2, and 3, all of which are highly expressed during development of the nervous system and are responsible for differentiation and survival of neurons.35,36 It has already been reported that chronic treatment of venlafaxine in patients with major depressive disorder increases expression of TGF-β1. However, unlike TGF-β1, the effect of antidepressants on the expression of TGF-β3 has hardly been studied. Recently, Wu et al.37 demonstrated that transient transduction of TGF-β3 by intra-amniotic gene transfer prevents cleft palate in Tgfβ3-/- mouse embryos. This finding suggests that TGF-β3 may be involved in embryonic development as well as development of the nervous system. Together with our observation that TGF-β3 was up-regulated by venlafaxine treatment during neural differentiation, the involvement of TGF-β3 in inhibiting cleft palate suggests that venlafaxine positively engages developmental processes via control of TGF-β3.
DPYSL3, also known as collapsing response mediator protein 4 (CRMP4), is involved in neuronal differentiation, neuronal regeneration, and outgrowth of neurites and axons.38,39 DPYSL3 is highly expressed in the adult brain, and its expression can be observed in new neurons during adult rat neurogenesis.40 In addition, CRMP2, one of CRMP family members, plays a pivotal role in outgrowth of axons and dendrites.41 In a study of Yoshimura et al.,42 the knockdown of CRMP2, a member of CRMP family, inhibited BDNF-induced axon elongation and branching, indicating that CRMP2 is associated with BDNF-induced axon outgrowth. As a result, CRMP family including CRMP2 and CRMP4 plays a pivotal role in growing axon of neurons. It was previously reported that the mRNA expression levels of TOAD-64, a member of the DPYSL3 family, are increased in the EC cell line P19 following exposure to RA during early neuronal differentiation.43 These findings suggest that the DPYSL3 family, including TOAD-64, may play an important role in neuronal differentiation and acquisition of a neuronal phenotype. In our study, the expression of DPYSL3 mRNA in cells undergoing differentiation and treated with venlafaxine for 1 day was significantly increased. The expression of DPYSL3 mRNA and protein after treatment with venlafaxine for 7 days was also increased compared with control cells; however, there was a noticeable difference between mRNA and protein expression compared with control cells. Together with previous studies, our data suggest that venlafaxine may affect neuronal differentiation by facilitating up-regulation of DPYSL3.
The expression levels of PKM mRNA and protein were up-regulated by treatment with venlafaxine after differentiation for 1 and 7 days. In glycolysis, PKM functions to generate pyruvate and glycolytic ATP to provide energy for living cells. Glucose metabolism in the brain not only is essential for normal function but also plays an important role in synaptic transmission.44 Ishida et al.45 have shown that PKM is responsible for transporting neurotransmitters such as GABA, dopamine, and serotonin into synaptic vesicles. In addition, they identified that PKM regulates the transfer of glutamate into synaptic vesicles through ATP consumption. Also, Glutamate and serotonin signaling are associated with synaptic plasticity, BDNF transcript and other neurotrophic factors.46 Bani-Yaghoub et al.47 showed that PKM is up-regulated during differentiation of the human teratocarcinoma line NT2/D1 into neurons, indicating that differentiation itself encourages the expression of proteins associated with metabolic mechanisms. In addition, Ryou et al.48 suggested that pyruvate has protective effects against ischemia-reperfusion brain injury. Consistent with these findings, we identified up-regulation of PKM by venlafaxine during neural differentiation. These results suggest that PKM is involved in pyruvate metabolism pathway, thus it may affect cellular differentiation and survival as well as events involved in neurotransmission.
The expression levels of P4HB mRNA and protein were significantly decreased after differentiation for 7 days and simultaneous treatment with venlafaxine. On the contrary, there was no difference between venlafaxine-treated cells and control cells after differentiation for 1 day. P4HB, known as protein disulfide isomerase (PDI), catalyzes the formation, isomerization, and breakage of disulfide linkages.49,50 P4HB has been reported to have opposing functions inside cells based on its concentration. Specifically, at high concentrations, P4HB suppresses aggregation of misfolded proteins, while at low concentrations, it promotes aggregation.49,50 On the other hand, a previous study showed that the toxicity of mutant huntingtin exon1 and Aβ peptides derived from amyloid precursor proteins could be inhibited by suppressing P4HB in rat brain cells.46 As suggested by others, this observation suggests the possibility that P4HB is a novel target for developing therapies for neurodegenerative disorders such as Huntington's disease and Alzheimer's disease.50,51,52 Based on previous reports49,50,51,52,53 and our own observation that chronic treatment of venlafaxine in vitro decreases expression of P4HB, additional studies associated with the cellular effects of antidepressants on the function of P4HB are warranted.
We found that treatment with venlafaxine resulted in decreased expression of not only HIP2 mRNA and protein after differentiation for 1 day, but also HIP2 mRNA after differentiation for 7 days. HIP2 is well known to interact with huntingtin, and affects the pathogenesis of both Huntington's and Alzheimer's disease.54,55 In addition, HIP2 promotes aggregate formation and cell death in polyglutamine-induced neurodegeneration.56 Likewise, Song et al.57 recently showed that knockdown of HIP2 in cortical neurons inhibits neuron death by endoplasmic reticulum stress. Taken together, the observed decrease in HIP2 expression by venlafaxine during neural differentiation suggests the possibility of venlafaxine as a drug candidate for treating neurodegenerative diseases in addition to depressive disorders.
Finally, we used MetaCore pathway analysis software (GeneGo Inc.) to identify top scoring networks that appeared to be involved in the effects of venlafaxine by analyzing differentially expressed proteins after neural differentiation for 1 and 7 days. The observed networks may offer useful information concerning physical connectivity and functional relationships among proteins that were differentially regulated by venlafaxine. Recently, rapid antidepressant action of ketamine has been focused on the mechanisms of novel treatment in depression.58,59 Mammalian target of rapamycin (mTOR) signaling pathway underlie rapid antidepressant effects. Yu et al.60 observed that venlafaxine prevented the effect of calcineurin inhibitor, which increased depressive-like behavior and reduced mTOR activity. In this study, we found that PKM and TGF-β regulated by venlafaxine were involved in activation of mTOR signaling pathway.61,62
In conclusion, we identified proteins that were differentially regulated by venlafaxine during neural differentiation of EBs derived from NCCIT cells. In particular, PKM was significantly up-regulated by venlafaxine during early RA induced-differentiation of EBs, while HIP2 was significantly down-regulated. Our findings suggest several proteins regulated by venlafaxine that are associated with cellular differentiation, survival of cells, cytoskeleton remodeling and metabolism of cell growth and proliferation. Also, these proteins are related with cytoskeleton remodeling pathway of neurofilament, pyruvate metabolism pathway and apoptosis and survival pathway.
Based on our findings, treatment with venlafaxine may affect neural differentiation in human embryonic carcinoma cells. Furthermore, these results will improve our understanding of the molecular mechanisms of venlafaxine.
Acknowledgments
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Korean Government (MSIP) (No. 2012-0009212, No. 2011-0030049).