A Functional Polymorphism in the DRD1 Gene, That Modulates Its Regulation by miR-504, Is Associated with Depressive Symptoms
Article information
Abstract
Objective
The aim of this study was to examine a possible association between depressive symptoms and a functional polymorphism (rs686) that modulates the regulation of DRD1 gene by miR-504.
Methods
A total of 239 young Colombian subjects were evaluated with the Patient Health Questionnaire-9 (PHQ-9) scale and genotyped for the rs686 polymorphism. A linear regression model, corrected by age and gender, was used.
Results
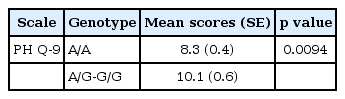
A significant association between the rs686 polymorphism and PHQ-9 scores was found, under a dominant genetic model (p=0.0094).
Conclusion
These results provide novel evidence about the growing role of inherited variants in binding sites for brain-expressed miRNAs on depressive symptomatology.
INTRODUCTION
Major depressive disorder (MDD) is one of the most common psychiatric disorders around the world, with an estimated overall point prevalence of around 4.7% and being a leading cause of disability worldwide [1]. MDD is a complex disorder caused by environmental and genetic factors, with an estimated heritability of 37% [2]. Although several genetic factors have been proposed as candidates for MDD, there is the need for the use of novel approaches, such as the analysis of novel functional polymorphisms in other genes involved in neural mechanisms and the study of endophenotypes, such as depressive symptoms [3].
Although polymorphisms in genes involved in the serotoninergic systems have been widely studied as possible risk factors for MDD, variations in genes involved in the dopaminergic systems have been proposed as novel candidates for MDD, taking into account the role of dopamine in neural mechanisms involved in motivation and reward [4]. Meta-analyses have found that two functional variants in genes involved in the serotoninergic and dopaminergic systems [encoding the serotonin transporter (SLC6A4) and the dopamine receptor 4 (DRD4)] are significantly associated with the susceptibility for MDD [5], highlighting the possibility that additional variants in other dopaminergic and serotoninergic genes might represent novel candidates for MDD.
The Dopamine receptor D1 (DRD1) gene is located on 5q35.1, has two exons and encodes the dopamine receptor 1, which has 446 amino acids. DRD1 is the most abundant dopaminergic receptor found in the central nervous system [6]. This protein is a member of the D1 subfamily of dopaminergic receptors, together with the dopamine receptor 5 (DRD5), which have seven-transmembrane domains and stimulate the adenylate cyclase pathway and the production of cAMP [6]. This gene has been involved in social cognition, attentional functioning, reinforcement mechanisms, executive functioning, working memory and neuropsychiatric disorders such as alcohol dependence and pathological gambling [7,8].
The only functional variant that has been found in the DRD1 gene, until now, is the rs686 single nucleotide polymorphism (SNP), which is an A/G polymorphism located in the 3’ untranslated region (3’ UTR) [9]. In a luciferase assay, it was found that this polymorphism leads to allele-specific effects on differential gene expression of the DRD1 gene, with the G allele showing a lower luciferase activity in comparison with the A allele (p<0.05) [9], which is due to the fact that this SNP is located in the binding region of miR-504 [10]. miR-504 is a miRNA expressed in cortical and hippocampal regions of the human brain [11], known to regulate the density of dendritic spines in cultured hippocampal neurons [11] and it has been shown that it is upregulated in post-mortem brains of bipolar disorder patients [12] and in a rat model of cocaine-induced conditioned place preference [13]. miRNAs are a recent category of non-coding RNAs that regulate the expression levels of a large number of proteincoding genes [14] and genetic variations in their binding sites represent interesting novel candidates for major psychiatric disorders [14], including MDD [15].
The functional rs686 polymorphism in DRD1 gene has not been previously associated with depressive symptomatology. The aim of the current study was to evaluate a possible association between a functional polymorphism regulated by miR-504 in the DRD1 gene and depressive symptoms (measured with the PHQ-9 scale).
METHODS
Participants
Two hundred thirty nine healthy Colombian individuals were included in the current study. The subjects were unrelated and had all four grandparents born in Colombia and participants with neurological or other major psychiatric illnesses, according to self-report, were excluded.
The study was approved by the Institutional Ethics Committee (Universidad Antonio Nariño; 07062015-1) and all participants signed a written informed consent.
Phenotypic evaluations
To assess depressive symptoms, the Patient Health Questionnaire-9 (PHQ-9) was used [16]. The PHQ-9 is a self-report scale designed to measure symptoms of depression based on the DSM-IV criteria and it has 9 items that evaluate the presence of depressive symptoms in the last 2 weeks. The recommended cut-off point is ≥10, indicative of depressive symptoms of clinical significance [16]. This instrument was selected because the PHQ-9 has demonstrated excellent reliability and validity in different languages, included Spanish [17]. Furthermore, this questionnaire has been used by several genetic studies for endophenotypes for MDD [18-20]. The Cronbach’s alpha for the PHQ-9 in the current sample was 0.84.
DNA genotyping
DNA extraction was carried out from 400 μL of peripheral blood using a salting out method [21] and DNA was normalized to 10 ng/μL. For the analysis of the functional polymorphism in the DRD1 gene (rs686), a TaqMan SNP genotyping assay (Applied Biosystems, Foster City, CA, USA, C_1011786_10) was used. It was carried out with 1X of TaqMan Genotyping Master Mix (Applied Biosystems), 1X of TaqMan Pre-Designed SNP Genotyping Assay (Applied Biosystems) and 20 ng of genomic DNA for a final volume of 10 μL. Fifty PCR cycles were run in a CFX96 Touch Real-Time PCR system (BioRad, Hercules, CA, USA) and the genotypes were analyzed using the CFX Manager software v.3.0 (BioRad).
Statistical analysis
Normal distribution from the PHQ-9 scores was examined through the skewness and kurtosis and the internal structure of the scale was evaluated through exploratory factorial analysis [22]. These statistical analyses were performed using the Statistical Package for the Social Sciences program (SPSS ver. 18; SPSS Inc., Chicago, IL, USA).
For the analysis of allele and genotype frequencies, Hardy Weinberg equilibrium and the association of the DRD1 polymorphism with PHQ-9 scores, the SNPStats program was used [23]. This program employs a linear regression model, which was adjusted for age and gender, in order to find a possible association between the SNP in the DRD1 gene and PHQ-9 scores, under a dominant genetic model. The best genetic model was selected based on the Akaike Information Criterion [23]. A p value< 0.05 was considered as significant.
RESULTS
The mean age of the participants was 21.3 years (SD= 4.0; range 18–57 years) and 76% were women. In our sample, the mean (SD) score for the PHQ-9 was 9.12 (5.33) and 42 subjects (17.1%) had PHQ-9 scores higher than 10.
The allelic frequencies for the A and G alleles were 0.74 and 0.26, respectively. We found the A/A genotype in 133 (56%), the A/G genotype in 88 (37%) and the G/G genotype in 18 (7%) of the 239 subjects analyzed. The genotype frequencies in our sample were in Hardy-Weinberg equilibrium (p=0.5).
We found a statistical significant association between the DRD1 polymorphism and the PHQ-9 score, under a dominant genetic model, using a linear regression model corrected by age and gender. The subjects with the A/G and G/G genotypes showed higher scores, in comparison to the A/A genotype, for depressive symptoms measured with the PHQ-9 scale (Table 1). We obtained mean scores (SD) for the PHQ-9 scale of 8.3, 10.2 and 9.6 for the carriers of the A/A, A/G, and G/G genotypes, respectively. An exploratory analysis of individual symptoms showed that items 1, 6, and 9 of the PHQ-9 (Anhedonia, Feeling bad about self and Thoughts of self-harm) had a significant association with the SNP in the DRD1 gene (p values of 0.03, 0.03, and 0.04, respectively).
DISCUSSION
Dopamine receptors play key roles in mediating dopamine actions in the central nervous system and they represent plausible candidate genes for genetic studies on MDD [6]. The DRD1 gene has been associated with the modulation of cognitive processes, particularly regarding its role within the prefrontal cortex [24] and in addition it has been involved in other brain processes such as reward and reinforcement mechanisms [25].
To date, the only polymorphism which has been functionally validated in the DRD1 gene is the rs686 polymorphism [10]. This polymorphism affects the expression of the DRD1 gene due to the fact that it is located in the binding site for miR-504, in the 3’UTR: it was observed that the G allele was associated with decreased expression of the DRD1 gene [10]. A recent study found that miR-504 expression is significantly associated with depressive behaviors in stressed rats and suggested that miR-504 may mediate the down-regulation of DRD1 and DRD2 expression in the nucleus accumbens [26].
Both alleles of rs686 polymorphism have been associated with different disease processes; the A allele has been associated with alcohol dependence [27] and the G allele has been associated with many diseases, including bipolar disorder, nicotine dependence, compulsive eating and pathological gambling [28]. Interestingly, previous studies have associated this polymorphism with the DRD1 activation in prefrontal cortex [10].
Our findings can be supported by previous reports that have linked the mesolimbic dopamine reward circuit with major depressive disorder [29]. This circuit has been widely correlated with drug reward as well as with natural rewards, such as food, sex, and social interactions, which can elicit dopamine release [30]. Abnormalities in these behavioral domains are evident in depression and other mood disorders; for example, subjects with MDD prominently exhibit loss of motivation and anhedonia and show abnormalities in several adaptive functions, such as appetite, sleep, and circadian rhythms [31]. Several individual symptoms assessed by the PHQ-9, such as Items 1 and 2 (Anhedonia and Feeling down), have been associated with function of the dopaminergic systems [32,33].
In the last decade, reward processing has been considered one of the most promising intermediate phenotypes for depression [34]. Changes in reward circuits are suggested as a core mechanism underlying depression pathophysiology [35]. Processing and integration of the reward are based on an interconnected dopamine-rich neural network, which includes the midbrain, amygdala, striatum, anterior cingulate cortex, orbitofrontal cortex, and medial prefrontal cortex [36]. Studies with functional MRI and positron emission tomography (PET) have shown that depressive behavior could be correlated with reduced rewardrelated activation in this network [37]. However, GWAS studies have not linked candidate genes for depression with reward processing dysfunction [38] with the only exception of the piccolo gene (PCLO) which has been associated with dopaminergic functioning [39].
Previous studies have analyzed other SNPs in the DRD1 gene in MDD patients. Koks et al. [40] evaluated the association of 91 SNPs in 21 candidate genes with MDD in Estonian patients, including other 7 SNPs in the DRD1 gene (located on the 5’UTR), and they did not find a significant association, after corrections for multiple testing, between these polymorphisms and MDD. Pearson-Fuhrhop et al. [41] explored the effect of dopamine genetic risk scores for polymorphisms in five genes involved in the dopaminergic system (including 1 SNP in the DRD1 gene, located on the 5’UTR) on depressive symptoms and in a subsample of healthy participants, analyzed with the CES-D scale, this association was significant. Corrales et al. analyzed 170 SNPs in 21 candidate dopaminergic and serotonergic genes (including other 5 SNPs in DRD1 gene, one of them located on the 5’UTR) in a sample of healthy children and adolescents from Costa Rica, assessed for depressive symptoms using the Child Depression Inventory (CDI) and none of these polymorphisms were significant [42].
It has been shown that the PHQ-9 is a valid instrument for measuring depression outcomes [43], since it is based on the criteria of the Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) for MDD. PHQ-9 can be used as a diagnostic assessment and it includes items that assess depressed mood or anhedonia [44].
In summary, we present the first report of an association between depressive symptoms and the functional rs686 polymorphism located in the DRD1 gene, which is known to modulate the regulation by miR-504 [10]. Our findings add evidence about the potential role of brain-expressed miRNAs in the pathophysiology and genetic risk of major depression [14,15]. The limitations of our study are the analysis of a single SNP in the DRD1 gene and use of a single instrument for analysis of depressive symptoms. Our sample size had a larger proportion of women, due to a higher participation rate of female subjects, as it has been shown in other genetic studies of behavioral endophenotypes [21,45].
Acknowledgements
This study was supported by a research grant from Colciencias (grant # 823-2015). We thank Andres Camargo, whom helped with recruitment and evaluation of subjects.