|
 |
- Search
| Psychiatry Investig > Volume 12(1); 2015 > Article |
Abstract
Objective
A relevant part of the social and personal burden caused by Bipolar Disorder (BD) is related to depressive phases. Authors investigated the genetic impact of a set of variations located in CRY1, a gene involved in the control of the circadian rhythms, towards depressive episodes in a sample of bipolar patients from the STEP-BD sample. As a secondary analysis, CYR1 variations were analyzed as predictors of sleep disruption.
Methods
654 bipolar patients were included in the analysis. Data were available genome-wide. The part of the genome coding for the CRY1 was imputed and pruned according to standards in the field. 7 SNPs were available for the analysis. A correction for multitesting was applied and we had sufficient power (0.80) to detect a small-medium effect size (0.22) between two allelic frequencies each one represented by at least 300 subjects.
Results
Intronic rs10861688 was associated with the number of depressive events corrected for the times patients were assessed during the period of observation. In particular, AA subjects (n=21) had 4.46±3.15 events, AG (n=141) had 3.08±3.17 and GG (n=342) 2.65±2.97 (p=0.0048, beta=-0.22). No other significant associations were reported.
Bipolar Disorder (BD) is a severe chronic disease with periods of remission and relapses. The hallmark of the disorder is a pathological mood swing between depressed and elated mood. Prevalence worldwide is estimated to be about 1%,1 which increases to 5% when broader diagnostic criteria are used.2 The typical onset occurs in early adulthood, but later onset may occur.3 An hypomanic or depressive episode is typically seen at the beginning of the disorder. A relevant part of the social and personal burden caused by BD is related to the depressive phases.3 Suicide is frequent among depressed bipolar patients4 and, along with that, BD ranked in the top list of the causes of non fatal burden, accounting for 2.5% of the total YLD (years lost for disability), in 2000 [http://www.who.int, and references therein]. Thus, the understanding of biological bases of BD, the definition of the most suitable treatments for each individual, and the identification of the individuals at risk of poor response to common treatments is a priority for the scientific community worldwide. Unfortunately, the pathogenesis of BD is not yet understood. BD has a heritability of 0.75 explained largely by common genetic variants,5 and it is thought to be the result of the interaction between genetic and environmental factors. There is solid evidence that stressful life experiences during early childhood may increase the risk of BD (nearly twofolds),6 and intervening stressors may thoroughly shape the prognosis of the disease.2 On the other hand, there is no clear cut consensus about which genes and variations may account for the risk of BD. A complete and excellent review on this topic can be found at.7 In particular, at least 12 independent GWAS studies recently reported findings that point to the genes and variations that may be at risk for BD, but the replication rate between them is very low,7 consistently with the genetic complexity of the disorder. An overview of the recent GWAS for BD can be found at [http://www.genome.gov/gwastudies/]. The biological counterpart of the interaction between the genetic predisposition and the intervening stressors is thought to be an imbalance in the neuronal interplay between key regions of the central nervous system including the prefrontal cortex, the amygdala, the anterior cingulate, the thalamus and the regions of the basal ganglia that are functionally related to them. In particular, BD would be the result of a deregulation of the synaptic function, the glutamatergic and GABAergic systems being primary involved. For a complete review on this topic, please refer at.2 Nevertheless, the glutamatergic and GABAergic systems are widespread in the brain and are activated at the common end of a variety of different biological processes. Thus, the identification of a set of genes that specifically drive the risk of BD still eludes the scientific efforts worldwide. A similar pattern of findings is retrieved when response to treatment is taken into consideration. Even though evidence is accruing showing a genetic impact towards response to common pharmacological treatments for BD (for some reviews about this topic please refer to),8,9,10,11 there is not a groundbreaking finding in the field to be translated into clinical practice, at the best of our knowledge. In the present work we analyzed a number of variations localized in CRY1 [Cryptochrome 1 (Photolyase-Like)], a gene involved in the circadian rhythm regulation,12 as modulator of depressive and manic relapses in a sample of 654 bipolar patients treated according to standard pharmacological guidelines. As a secondary analysis, we investigated the impact of these variations towards sleep disruption. The disruption of circadian rhythms is an hallmark of BD,13 and sleep disturbance may precede the clinical onset of the disease, is a residual symptom during euthymic periods and may be associated with the cognitive impairment that characterizes the disease.14 Consistently with that, there is convincing evidence that the genes that regulate the circadian rhythms are related to response to pharmacological treatments of BD15 and susceptibility to it (reviewed in 16). CRY1 encodes a flavin adenine dinucleotide-binding protein that is a key component of the circadian core oscillator complex. The protein is widely conserved across plants and animals, which suggests that variations located in its coding frame are particularly disruptive. There is evidence that some genetic variations harbored by CRY1 are associated with mood disorders. Rs2287161 was found to be associated with mood disorder (combined unipolar and bipolar) in a sample of 335 patients with unipolar disorder, 199 patients with bipolar disorder and 440 matched controls.17 Another variant located in CRY1 (rs8192440) was found to be associated with good response to lithium response in a sample of 282 bipolar patients.18 These genetic findings are replicated in the present work in a larger sample of bipolar sample treated in a naturalistic setting.
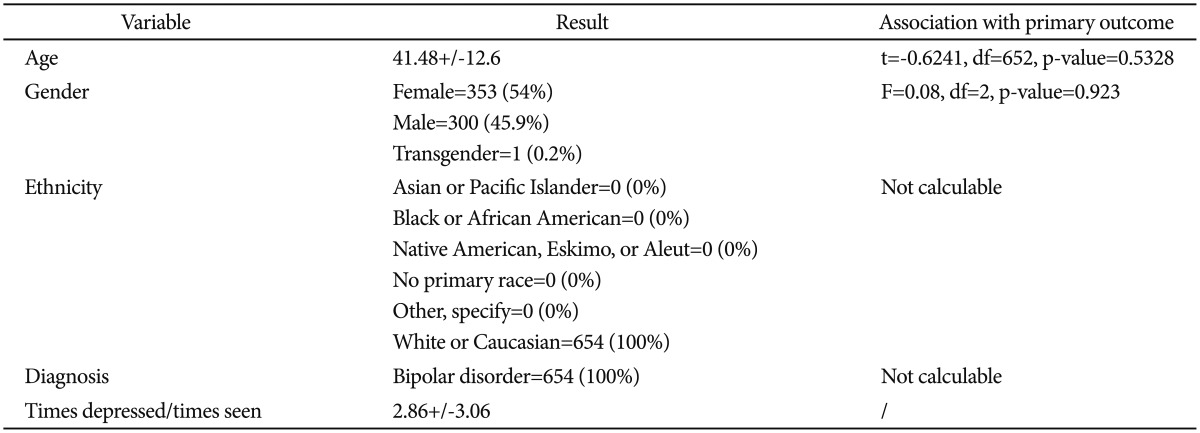
The sample under investigation was retrieved from the public available STEP-BD protocol.19 During the study, bipolar patients of every subtype with age ≥15 years are accessioned into a study registry. All patients signed a written informed consent before entering the study. All patients receive a systematic assessment battery at entry and are treated by a psychiatrist (trained to deliver care and measure outcomes in patients with bipolar disorder) using a series of model practice procedures consistent with expert recommendations. At every follow-up visit, the treating psychiatrist completes a standardized assessment and assigns an operationalized clinical status based on DSM-IV criteria. Patients have independent evaluations at regular intervals throughout the study and remain under the care of the same treating psychiatrist while making transitions between randomized care studies and the standard care treatment pathways. In the present study we only considered records from the naturalistic first phase of the study. Table 1 reports the characteristics of the sample under analysis.
The ratio between the times patients were found to be definitively depressed and the times patients were observed during the phase of the study under analysis was the outcome of choice. This phenotype was designed to include all the observed depressed phases, in order to include as many patients as possible to increase the power of the study. Care was taken to control for a possible clinical bias: more severe patients might have been seen more often compared to less severe ones. In order to do so, the correlations between the phenotype of choice and a set of other phenotypes calculated at standard timepoints (number of depressive phases from 30-90-120 and so forth days from the beginning of the study)-a more classical approach to this kind of studies-were calculated. We had confirmation that the phenotype under analyses significantly correlated with almost all the phenotypes at different timepoints, with the advantage of having 0% of missing values. The only timepoint at which the correlation was not significant was after 30 days from the beginning of the study. Nevertheless, the number of missing information for this timepoint (94%) may be held accountable for the lack of association.
The sociodemographic and clinical variables were investigated as possible stratification factors. A story of treatment for alcohol related problems resulted to be significantly associated with the outcome (F=2.54, df=4, p-value=0.038) as was included in the analysis as a covariate. Treatment (combination of treatments) correlated with the number of depressive episodes during the period of observation (combination of antipsychotics and antidepressants throughout the study F=18.28; p=2.07e-05; combination of mood stabilizers and antidepressants during the study F=39.82; p=4.07e-10; combination of mood stabilizers and antipsychotics during the study F=26.72; p=2.8e-07; combination of mood stabilizers, antipsychotics and antidepressants during the study F=28.2; p=1.33e-07) and was included in the analysis as covariate. The administration of antidepressant alone did not correlate with the number of depressive events (F=2.019; p=0.156), consistent with literature and guidelines in the field.
Age at the first depression episode negatively correlated with the secondary outcome (t=-3.2, df=638, p-value=0.001, r=-0.12) and was included in the analysis as a covariate. An history of legal problems was significantly associated with sleep disruption (sleep disruption frequency in subjects with legal problems was 2.35±1.8, and 1.81±1.71 in subjects without a story of legal problems) (F=4.7, df=2, p=0.008). This latter variable was not included in the analysis because it was classified as a psychopathological stratification factor.
We had sufficient power (0.80) to detect a small-medium effect size (0.22) between two allelic frequencies each one represented by at least 300 subjects (R-cran20 "pwr" package served for the analysis).
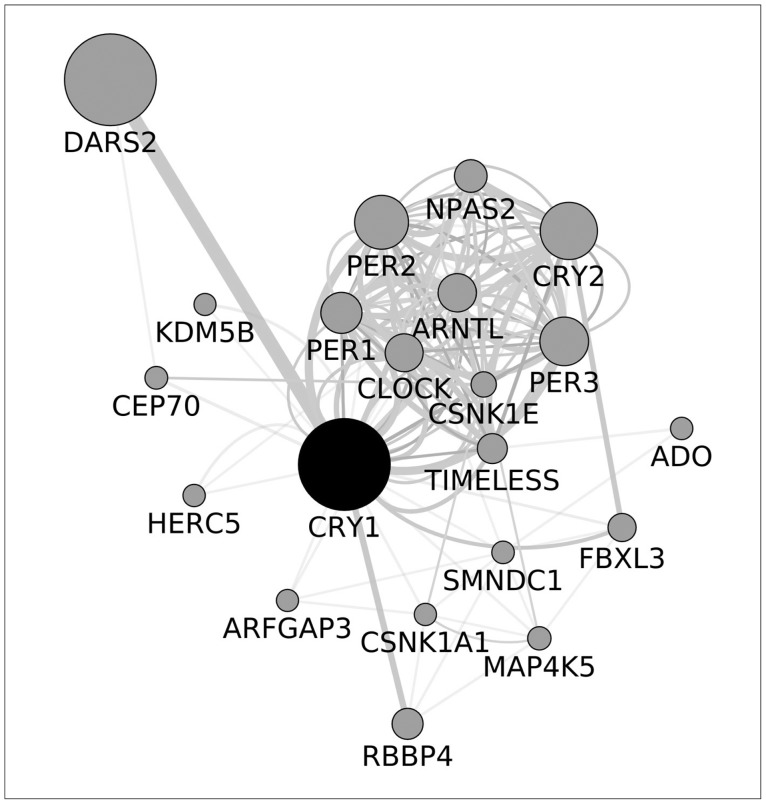
CRY1 was the candidate gene for the analysis. Variations located in its genomic frame were imputed and pruned, please refer to the following paragraph. Moreover, the entire CYR1 molecular pathway was investigated as a secondary analysis. Candidates were selected according to the Cytoskape (http://www.cytoscape.org/) analysis of the CRY1 molecular pathway (Figure 1).
Imputation was run for the genes that belong to the pathway under analysis in order to decrease the computational effort. The CEU HapMap 1000 genomes served for the analysis. From the original (not pruned) set of 11 SNPs harbored by CRY1 we obtained 7 SNPs that passed the imputation quality control (info>0.9) and pruning (r2>0.2). Pruning was undertaken after imputation.
The p value for a significant result was set at 0.05/7=0.007 (Bonferroni correction). 7 was the number of SNPs available for analysis after imputation and pruning.
Covariated linear regression was the statistical model for the analysis. Plink served for the analysis.21 For the secondary genetic analysis a pathway enrichment analysis was undertaken as follows. The genomic sections that contain the genes included in the selected pathways were identified through PLINK annotation. These sections were imputed, checked for quality (info>0.9) and pruned (r2>0.2), please refer to previous paragraphs, and their association with the phenotype (schizophrenic patients vs controls or bipolar patients vs. controls) tested. From the remaining part of the genome a random selection of variations was taken, their number summing up to the number of analyzed SNPs within the pathway of interest, and all of them belonging to genes randomly selected throughout the genome. The association of this random list of variations was tested against the phenotype under analysis. Then, the prevalence of variations significantly (p<0.01) associated with the outcome in the pathway under analysis and the random pathway were tested for a significant different distribution (Fisher exact test). This operation was repeated 106 times (permutation) selecting different groups of random SNPs as a control group, under the same parameters described above. 106 Fisher Tests where then conducted and the permuted p was extracted according to Phipson.22 The whole pathway and subpathways were tested, and the genes that held the highest number of variations (at least ten times the expected values) associated with the phenotype were used to define a further sub-pathway.
We had evidence for an association between rs10861688 - located in an intronic region in CRY1-and the number of depressive events corrected for the times patients were assessed during the period of observation. In particular, AA subjects (n=21) had 4.46±3.15 events, AG (n=141) had 3.08±3.17 and GG (n=342) 2.65±2.97 (p=0.0048, beta=-0.22). The entire molecular pathway was not found to be significantly enriched after the permutation analysis.
CRY1 did not harbor variations significantly associated with the sleep disruptions, nor the CRY1 pathway was found to be enriched in bipolar patients presenting with sleep disruption. Within the pathway a trend for a significant association was found for rs707472 (p=0.006, beta=0.27) harbored by PER3. Rs10861688 harbored by PER3 displayed a trend of association with sleep disruption (p=0.47, beta=0.07), along with two other SNPs located in that gene (rs6539299 and rs7303842 respectively p=0.07, beta=-0.6 and p=0.08, beta=-0.06).
Many common psychiatric conditions are associated with disrupted sleep, indicative of imbalanced circadian timing in these disorders. Circadian rhythm hypotheses have proposed for the explanation of major depressive disorder (MDD) and bipolar disorder (BD), as many data support the association between circadian rhythm dysfunction and mood disorder (reviewed by 23,24,25). Various phases of mood disorder (depressed, manic, episode prodrome, and interepisode periods) are associated with circadian rhythm abnormalities and the depressive symptom severity correlates with disturbed sleep-wake and circadian rhythm. Patients in the manic phase generally have a decreased need for sleep, while insomnia and hypersomnia are both commonly found in depressive patients. Sleep disturbance is a common prodromal symptom of both mania and depression. In fact, it seems to be a predisposition for circadian rhythm alteration and the related sleep disturbance in mood disorder patients. Moreover, sleep disturbances and instability of circadian rhythm continue when mood disorder patients are not acutely ill.14,26 Since circadian rhythms disruption has been found among patients with major depressive disorder (MDD) and BD, it has been hypothesized that abnormalities in the molecular clock could underlie the development of mood disorders. Circadian gene polymorphisms seem to be associated with both mood disorders and alterations in circadian rhythm, therefore certain circadian gene variants may represent susceptibility factors for mood spectrum disorders. Such connections between polymorphisms in circadian genes such as CLOCK,27 ARNTL1,28 NPAS2,29 PER2,28,29 PER330 and TIMELESS31,32 have been demonstrated for bipolar disorders, and to a lesser extent for recurrent depressive disorders and seasonal affective disorders.29,33 More recently, the correlation between genes involved in circadian clock and depression has been further demonstrated by studying the expression of these genes in different regions of postmortem brains from control and MDD patients, thus showing alterations in canonical clock genes, such as ARNTL, PER1-2-3, NR1D1, DBP and DEC1-2.32 Here we focus our attention on the gene CRY1. Cryptochromes are blue-light absorbing photopigmenst that have been proposed to act as a photoreceptor for a variety of nonvisual light-responsive tasks. Cryptochrome proteins (CRY1 and CRY2 in humans and Cry1 and Cry2 in mouse) are codified by two genes with high degree of sequence homology to photolyase/plant blue-light photoreceptor gene family. Both genes are expressed throughout the body, but both they are expressed at high levels in the ganglion cells and the inner nuclear layer of the retina, which are known to be important for circadian photoreception. Moreover, CRY1 is expressed with a robust circadian rhythm in the suprachiasmatic nucleus (SCN).34 These pigments function as photoreceptors for setting the circadian clock;35 in fact, the circadian clock is maintained by an autoregulatory system involving 'clock' genes in the SCN.36 This system involves the interaction among positive and negative transcriptional/posttranslational regulators, including the transcription factors CLOCK and BMAL1, which drive the rhythmic transcription of three Period genes (PER1-3) and two Cryptochrome genes (CRY1, 2).37 These clock genes play different roles in retinal and suprachiasmatic nucleus circadian oscillators, having the retinal clock a pattern of gene dependence similar, but more severe than the SCN clock, with divergent gene regulation of rhythmic period.38 Molecular clocks do not oscillate with an exact 24-hour rhythmicity, but they are entrained to solar day/night rhythms by light. The mammalian proteins Cry1 and Cry2 are involved in the light control of the biological clock. Interestingly, light affects behavior in two ways: indirectly through the circadian timing system; directly through mechanisms that are independent of the circadian system. Both effects have been studied in regard to mood, thus suggesting that light may exert its antidepressant effect through a modulation of the homeostatic process of sleep.39 Concerning the role of CRY1 and 2 in clock regulation and, therefore, their possible role in depression and/or sleep disturbance, genetic knock-out mice for each Cryptochrome gene have been developed. The double knock-out mouse shows a defective biological clock.40 In particular, the Cry1-/- mice had a circadian period 1 hr shorter than wild type and the Cry1-/-/Cry2-/- mice were arrhythmic. Circadian clock controls many physiological processes, from body temperature to the sleep-wake cycle. In particular, there is a specific daily sleep-wake distribution during the 24 h that is a consequence of a signal from the circadian clock in SCN. In fact, in Cry1,2-/- mice there was a pronounced decrease in sleep time compared to wild-time animals and an increase of non-REM sleep proportion during the dark period.41
Circadian clock gene variants, including CRY1, have been previously associated with depression. In particular, the genetic variants of CRY1 (rs2287161), as well as of RORA (rs2028122), have been associate with depressive disorder, while those of RORB (rs7022435, rs3750420, rs1157358, rs3903529) and NR1D1 (rs2314339) with bipolar disorder, and those of NPAS2 (rs11541353) and CRY2 (rs10838524) with seasonal affective disorder or winter depression.42 Moreover, in a sample of patients with unipolar major mood depression (MDD) and with bipolar disorder (BD) statistically significant associations were found in 15 circadian genes, including CRY. In particular, there were identified significant associations in CRY1 (rs2287161), NPAS2 (rs11123857) and VIPR2 (rs885861) genes with the combined sample (MDDD+BD), whereas CLOCK (rs10462028) and VIP (rs17083008) were specifically associated in the BD, thus supporting the contribution of the circadian system to the genetic susceptibility to depression and suggesting that different circadian genes may have specific effects on depression polarity. Contrasting results have been obtained with no association between the clock gene Cryptochrome 1 and bipolar disorder.43 Here we have demonstrated the specific association between the presence of AA and, to a lesser extent AG, at rs10861688-located in an intronic region in CRY1-and a higher number of depressive events, while there was no correlation in bipolar patients presenting with sleep disruption. On the contrary, significant association with sleep disruption was found for PER3 allelic variants. Interestingly, polymorphisms in PER3 gene have been previously associated with the postpartum onset of bipolar disorder30 and with change of sleep patterns in females.44
In conclusion, the data here reported further support the role of genes controlling circadian rhythm in bipolar depression and sleep disturbance, demonstrating the need to deeply study the genetic variants of all clock genes in the different aspects of these psychiatric diseases, from the identification of new genetic markers to predict the risk of mood disorders to the opportunity of new safe and inexpensive potential approaches to treat these disorders.
Acknowledgments
Data and biomaterials were collected for the Systematic Treatment Enhancement Program for Bipolar Disorder (STEP-BD), a multi-center, longitudinal (5-8 years) project selected from responses to RFP #NIMH-98-DS-0001, "Treatment for Bipolar Disorder." The project was led by Gary Sachs, M.D., and coordinated by Massachusetts General Hospital in Boston, MA. The NIMH grant number was 2N01MH080001-001. Given the major public health implications of identifying genes responsible for severe neuropsychiatric disorders, the National Institute of Mental Health (NIMH) has funded a Human Genetics Initiative. The goal of this Initiative is to establish a national resource of clinical data and biomaterials that are collected from individuals with Alzheimer disease, schizophrenia, or bipolar I disorder (BP), in order to aid researchers in understanding the genetic bases of these disorders. The NIMH Bipolar Disorder Genetics Initiative is supported by the Office of Human Genetics & Genomic Resources in NIMH's Division of Neuroscience and Basic Behavioral Science (DNBBS). Since 1996, data and biomaterials (cell lines and DNA samples) have been available to qualified investigators who study the genetics of BP, and may be accessed by following a set of instructions. We gratefully thank Samuel Mexcur for his help in editing the final version of the article.
References
1. Ferrari AJ, Baxter AJ, Whiteford HA. A systematic review of the global distribution and availability of prevalence data for bipolar disorder. J Affect Disord 2011;134:1-13. PMID: 21131055.


2. Martinowich K, Schloesser RJ, Manji HK. Bipolar disorder: from genes to behavior pathways. J Clin Invest 2009;119:726-736. PMID: 19339764.



3. Goodwin GM, Anderson I, Arango C, Bowden CL, Henry C, Mitchell PB, et al. ECNP consensus meeting. Bipolar depression. Nice, March 2007. Eur Neuropsychopharmacol 2008;18:535-549. PMID: 18501566.


4. Clements C, Morriss R, Jones S, Peters S, Roberts C, Kapur N. Suicide in bipolar disorder in a national English sample, 1996-2009: frequency, trends and characteristics. Psychol Med 2013;43:2593-2602. PMID: 23510515.


5. Sullivan PF, Daly MJ, O'Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet 2012;13:537-551. PMID: 22777127.



6. Etain B, Henry C, Bellivier F, Mathieu F, Leboyer M. Beyond genetics: childhood affective trauma in bipolar disorder. Bipolar Disord 2008;10:867-876. PMID: 19594502.


8. Cruceanu C, Alda M, Rouleau G, Turecki G. Response to treatment in bipolar disorder. Curr Opin Psychiatry 2011;24:24-28. PMID: 21088584.



9. McCarthy MJ, Leckband SG, Kelsoe JR. Pharmacogenetics of lithium response in bipolar disorder. Pharmacogenomics 2010;11:1439-1465. PMID: 21047205.


10. Smith DJ, Evans R, Craddock N. Predicting response to lithium in bipolar disorder: a critical review of pharmacogenetic studies. J Ment Health 2010;19:142-156. PMID: 20433322.


11. Dallaspezia S, Benedetti F. Melatonin, circadian rhythms, and the clock genes in bipolar disorder. Curr Psychiatry Rep 2009;11:488-493. PMID: 19909672.


12. van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 1999;398:627-630. PMID: 10217146.


13. Westrich L, Sprouse J. Circadian rhythm dysregulation in bipolar disorder. Curr Opin Investig Drugs 2010;11:779-787.

14. Boland EM, Alloy LB. Sleep disturbance and cognitive deficits in bipolar disorder: toward an integrated examination of disorder maintenance and functional impairment. Clin Psychol Rev 2013;33:33-44. PMID: 23123569.


15. McCarthy MJ, Nievergelt CM, Kelsoe JR, Welsh DK. A survey of genomic studies supports association of circadian clock genes with bipolar disorder spectrum illnesses and lithium response. PLoS One 2012;7:e32091PMID: 22384149.



16. Mendlewicz J. Disruption of the circadian timing systems: molecular mechanisms in mood disorders. CNS Drugs 2009;23(Suppl 2):15-26. PMID: 19708722.


17. Soria V, Martínez-Amorós E, Escaramís G, Valero J, Pérez-Egea R, García C, et al. Differential association of circadian genes with mood disorders: CRY1 and NPAS2 are associated with unipolar major depression and CLOCK and VIP with bipolar disorder. Neuropsychopharmacology 2010;35:1279-1289. PMID: 20072116.



18. McCarthy MJ, Nievergelt CM, Shekhtman T, Kripke DF, Welsh DK, Kelsoe JR. Functional genetic variation in the Rev-Erbα pathway and lithium response in the treatment of bipolar disorder. Genes Brain Behav 2011;10:852-861. PMID: 21781277.



19. Sachs GS, Thase ME, Otto MW, Bauer M, Miklowitz D, Wisniewski SR, et al. Rationale, design, and methods of the systematic treatment enhancement program for bipolar disorder (STEP-BD). Biol Psychiatry 2003;53:1028-1042. PMID: 12788248.


20. A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing R Development Core Team; 2012,Available at: http://www.R-project.org.
21. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559-575. PMID: 17701901.



22. Phipson B, Smyth GK. Permutation P-values should never be zero: calculating exact P-values when permutations are randomly drawn. Stat Appl Genet Mol Biol 2010;9:Article39PMID: 21044043.


23. Courtet P, Olié E. Circadian dimension and severity of depression. Eur Neuropsychopharmacol 2012;22(Suppl 3):S476-S481. PMID: 22959112.


24. Coogan AN, Thome J. Chronotherapeutics and psychiatry: setting the clock to relieve the symptoms. World J Biol Psychiatry 2011;12(Suppl 1):40-43. PMID: 21905994.


25. Harvey AG. Sleep and circadian functioning: critical mechanisms in the mood disorders? Annu Rev Clin Psychol 2011;7:297-319. PMID: 21166537.


26. Plante DT, Winkelman JW. Sleep disturbance in bipolar disorder: therapeutic implications. Am J Psychiatry 2008;165:830-843. PMID: 18483132.


27. Lee KY, Song JY, Kim SH, Kim SC, Joo E-J, Ahn YM, et al. Association between CLOCK 3111T/C and preferred circadian phase in Korean patients with bipolar disorder. Prog Neuropsychopharmacol Biol Psychiatry 2010;34:1196-1201. PMID: 20600471.


28. Partonen T, Treutlein J, Alpman A, Frank J, Johansson C, Depner M, et al. Three circadian clock genes Per2, Arntl, and Npas2 contribute to winter depression. Ann Med 2007;39:229-238. PMID: 17457720.


29. Johansson C, Willeit M, Smedh C, Ekholm J, Paunio T, Kieseppä T, et al. Circadian clock-related polymorphisms in seasonal affective disorder and their relevance to diurnal preference. Neuropsychopharmacology 2003;28:734-739. PMID: 12655319.


30. Dallaspezia S, Lorenzi C, Pirovano A, Colombo C, Smeraldi E, Benedetti F. Circadian clock gene Per3 variants influence the postpartum onset of bipolar disorder. Eur Psychiatry 2011;26:138-140. PMID: 21316201.


31. Utge SJ, Soronen P, Loukola A, Kronholm E, Ollila HM, Pirkola S, et al. Systematic analysis of circadian genes in a population-based sample reveals association of TIMELESS with depression and sleep disturbance. PLoS One 2010;5:e9259PMID: 20174623.



32. Li JZ, Bunney BG, Meng F, Hagenauer MH, Walsh DM, Vawter MP, et al. Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc Natl Acad Sci U S A 2013;110:9950-9955. PMID: 23671070.



33. Etain B, Milhiet V, Bellivier F, Leboyer M. Genetics of circadian rhythms and mood spectrum disorders. Eur Neuropsychopharmacol 2011;21(Suppl 4):S676-S682. PMID: 21835597.


34. Miyamoto Y, Sancar A. Vitamin B2-based blue-light photoreceptors in the retinohypothalamic tract as the photoactive pigments for setting the circadian clock in mammals. Proc Natl Acad Sci U S A 1998;95:6097-6102. PMID: 9600923.



35. Thresher RJ, Vitaterna MH, Miyamoto Y, Kazantsev A, Hsu DS, Petit C, et al. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science 1998;282:1490-1494. PMID: 9822380.


36. Dunlap JC. Common threads in eukaryotic circadian systems. Curr Opin Genet Dev 1998;8:400-406. PMID: 9729714.


37. Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol 2001;63:647-676. PMID: 11181971.


38. Ruan GX, Gamble KL, Risner ML, Young LA, McMahon DG. Divergent roles of clock genes in retinal and suprachiasmatic nucleus circadian oscillators. PLoS One 2012;7:e38985PMID: 22701739.



39. Stephenson KM, Schroder CM, Bertschy G, Bourgin P. Complex interaction of circadian and non-circadian effects of light on mood: shedding new light on an old story. Sleep Med Rev 2012;16:445-454. PMID: 22244990.


40. Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci U S A 1999;96:12114-12119. PMID: 10518585.



41. Wisor JP, O'Hara BF, Terao A, Selby CP, Kilduff TS, Sancar A, et al. A role for cryptochromes in sleep regulation. BMC Neurosci 2002;3:20PMID: 12495442.



42. Partonen T. Clock gene variants in mood and anxiety disorders. J Neural Transm 2012;119:1133-1145. PMID: 22538398.


43. Nievergelt CM, Kripke DF, Remick RA, Sadovnick AD, McElroy SL, Keck PE Jr, et al. Examination of the clock gene Cryptochrome 1 in bipolar disorder: mutational analysis and absence of evidence for linkage or association. Psychiatr Genet 2005;15:45-52. PMID: 15722957.


44. Antypa N, Mandelli L, Nearchou FA, Vaiopoulos C, Stefanis CN, Serretti A, et al. The 3111T/C polymorphism interacts with stressful life events to influence patterns of sleep in females. Chronobiol Int 2012;29:891-897. PMID: 22823872.


Figure 1
Refer to the attached file. DARS2: aspartyl-tRNA synthetase 2, NPAS2: neuronal PAS domain protein 2, PER2: period circadian clock 2, CRY2: cryptochrome circadian clock 2, ARNTL: aryl hydrocarbon receptor nuclear translocator-like, PER1: period circadian clock 1, CLOCK: clock circadian regulator, PER3: period circadian clock 3, KDM5B: lysine (K)-specific demethylase 5B, CEP70: centrosomal protein 70 kDa, CSNK1E: casein kinase 1, epsilon, TIMELESS: timeless circadian clock, ADO: 2-aminoethanethiol (cysteamine) dioxygenase, HERC5: HECT and RLD domain containing E3 ubiquitin protein ligase 5, SMNDC1: survival motor neuron domain containing 1, FBXL3: F-box and leucine-rich repeat protein 3, ARFGAP3: ADP-ribosylation factor GTPase activating protein 3, CSNK1A1: casein kinase 1, alpha 1, MAP4K5: mitogen-activated protein kinase kinase kinase kinase 5, RBBP4: retinoblastoma binding protein 4.