 |
 |
- Search
| Psychiatry Investig > Volume 20(8); 2023 > Article |
|
Abstract
Objective
Methods
Results
Notes
Availability of Data and Material
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors have no potential conflicts of interest to disclose.
Author Contributions
Conceptualization: Yiwei Lin, Haimei Li, Lu Liu, Qiujin Qian. Data curation: Yiwei Lin, Haimei Li, Lu Liu. Formal analysis: Yiwei Lin, Haimei Li, Qiujin Qian. Funding acquisition: Haimei Li, Lu Liu, Qiujin Qian. Methodology: Yiwei Lin, Lu Liu. WritingŌĆöoriginal draft: Yiwei Lin, Qiujin Qian. WritingŌĆöreview and editing: all authors.
Funding Statement
This study was partly funded by the National Basic Research Development Program of China (grant number 973 program 2014CB846104), National Natural Sciences Foundation of China (grant numbers 81071109; 81301171).
ACKNOWLEDGEMENTS
Figure┬Ā1.
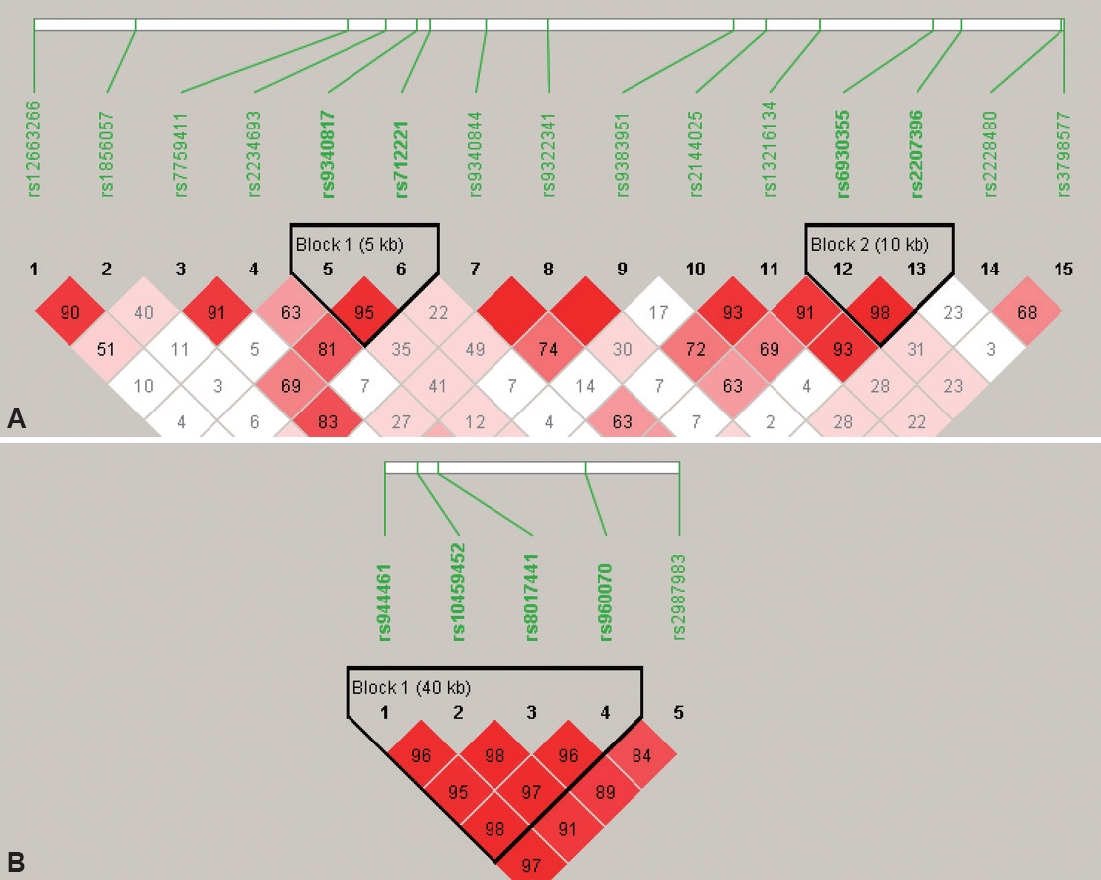
Figure┬Ā2.

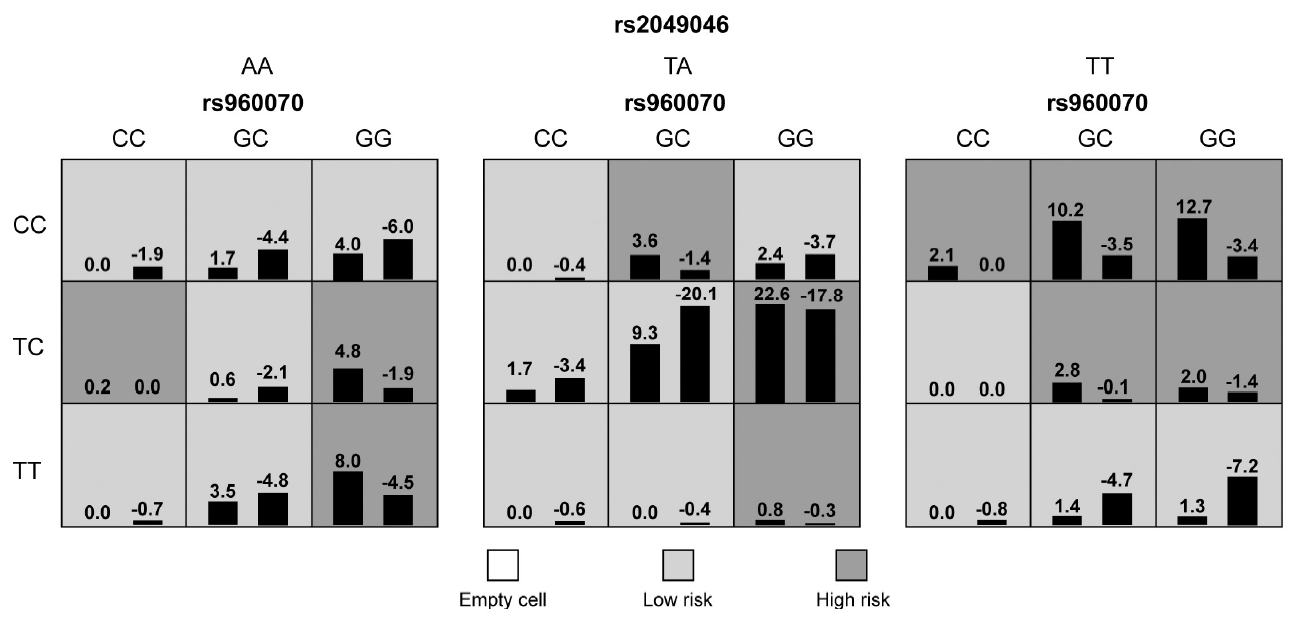
Figure┬Ā3.
Table┬Ā1.
Table┬Ā2.
Table┬Ā3.
| Gene | SNP* |
Alleles |
Genotypes |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Allele |
Ratio counts case, control | Frequencies case, control | Assoc. allele | Nominal p | Empirical p |
11 vs. 12+22 |
11+12 vs. 22 |
||||||
| 1 | 2 | OR (95% CI) | p | OR (95% CI) | p | ||||||||
| All subjects | |||||||||||||
| ESR1 | rs9340817 | A | C | 532:1522, 443:1467 | 0.259, 0.232 | A | 0.0480 | 0.4728 | |||||
| rs2228480 | A | G | 482:1580, 395:1523 | 0.234, 0.206 | A | 0.0344 | 0.3682 | ||||||
| ESR2 | rs10459452 | A | G | 1953:117, 1778:146 | 0.943, 0.924 | A | 0.0137 | 0.0554 | |||||
| rs8017441 | A | C | 1899:167, 1711:199 | 0.919, 0.896 | A | 0.0109 | 0.0426 | 1.480 (1.049-2.088) | 0.026 | 0.947 (0.24-3.65) | 0.937 | ||
| rs960070 | C | G | 1588:480, 1394:528 | 0.768, 0.725 | G | 0.0020 | 0.0076 | 0.656 (0.423-1.018) | 0.060 | 0.753 (0.61-0.93) | 0.008 | ||
| rs2987983 | A | G | 1426:644, 1267:657 | 0.689, 0.659 | A | 0.0408 | 0.1422 | ||||||
| Male | |||||||||||||
| ESR1 | rs2234693 | C | T | 687:1053, 426:784 | 0.395, 0.352 | C | 0.0184 | 0.2132 | |||||
| rs9340817 | A | C | 451:1279, 257:951 | 0.261, 0.213 | A | 0.0028 | 0.0344 | 1.415 (0.802-2.496) | 0.231 | 1.336 (1.04-1.72) | 0.024 | ||
| rs712221 | A | T | 814:926, 509:697 | 0.468, 0.422 | T | 0.0141 | 0.1666 | ||||||
| rs2228480 | A | G | 408:1332, 237:973 | 0.234, 0.196 | A | 0.0126 | 0.1470 | ||||||
| ESR2 | rs8017441 | A | G | 1603:141, 1083:125 | 0.919, 0.897 | A | 0.0348 | 0.1326 | |||||
| Female | |||||||||||||
| ESR1 | rs6930355 | C | T | 125:191, 224:457 | 0.396, 0.328 | C | 0.0372 | 0.3786 | |||||
| ESR2 | rs960070 | C | G | 253:71, 504:206 | 0.781, 0.710 | G | 0.0168 | 0.0604 | |||||
Table┬Ā4.
| Haplotypes | Block | Freq. | Case, control frequencies | Nominal p | Empirical p | ||
|---|---|---|---|---|---|---|---|
| All subjects | |||||||
| ESR1 | |||||||
| rs9340817-rs712221 | CA | 0.540 | 0.527, 0.554 | 0.0800 | 0.3166 | ||
| AT | 0.239 | 0.253, 0.224 | 0.0305* | 0.1294 | |||
| CT | 0.215 | 0.215, 0.214 | 0.9570 | >0.9999 | |||
| rs6930355-rs2207396 | TG | 0.434 | 0.438, 0.430 | 0.5991 | 0.9888 | ||
| CG | 0.346 | 0.352, 0.339 | 0.3767 | 0.8978 | |||
| TA | 0.220 | 0.210, 0.231 | 0.1003 | 0.3890 | |||
| ESR2 | |||||||
| rs944461-rs10459452-rs8017441-rs960070 | TGGC | 0.065 | 0.055, 0.075 | 0.0093 | 0.0268* | ||
| TAAC | 0.162 | 0.153, 0.171 | 0.1290 | 0.3924 | |||
| TAAG | 0.445 | 0.456, 0.433 | 0.1313 | 0.3958 | |||
| CAAG | 0.301 | 0.310, 0.292 | 0.2194 | 0.6274 | |||
| TAGC | 0.025 | 0.024, 0.028 | 0.4184 | 0.9020 | |||
| Male | |||||||
| ESR1 | |||||||
| rs9340817-rs712221 | CA | 0.544 | 0.527, 0.568 | 0.0244* | 0.0890 | ||
| AT | 0.233 | 0.254, 0.203 | 0.0012 | 0.0058* | |||
| CT | 0.216 | 0.214, 0.219 | 0.7265 | 0.9974 | |||
| rs6930355-rs2207396 | TG | 0.430 | 0.442, 0.413 | 0.1082 | 0.4002 | ||
| CG | 0.345 | 0.345, 0.345 | 0.9862 | >0.9999 | |||
| TA | 0.225 | 0.213, 0.243 | 0.0596 | 0.2240 | |||
| ESR2 | |||||||
| rs944461-rs10459452-rs8017441-rs960070 | TGGC | 0.062 | 0.055, 0.073 | 0.0535 | 0.1598 | ||
| TAAG | 0.445 | 0.455, 0.431 | 0.1901 | 0.5596 | |||
| TAGC | 0.025 | 0.023, 0.028 | 0.3840 | 0.8446 | |||
| TAAC | 0.158 | 0.155, 0.163 | 0.5993 | 0.9812 | |||
| CAAG | 0.306 | 0.309, 0.302 | 0.6825 | 0.9938 | |||
| Female | |||||||
| ESR1 | |||||||
| rs9340817-rs712221 | CA | 0.530 | 0.527, 0.531 | 0.9128 | >0.9999 | ||
| AT | 0.256 | 0.246, 0.260 | 0.6396 | 0.9916 | |||
| CT | 0.211 | 0.223, 0.206 | 0.5287 | 0.9718 | |||
| rs6930355-rs2207396 | TG | 0.445 | 0.415, 0.459 | 0.1926 | 0.5770 | ||
| CG | 0.348 | 0.393, 0.327 | 0.0395* | 0.1806 | |||
| TA | 0.206 | 0.415, 0.459 | 0.1926 | 0.5770 | |||
| ESR2 | |||||||
| rs944461-rs10459452-rs8017441-rs960070 | TAAC | 0.170 | 0.139, 0.184 | 0.0723 | 0.2382 | ||
| TGGC | 0.071 | 0.052, 0.079 | 0.1242 | 0.3828 | |||
| CAAG | 0.288 | 0.318, 0.274 | 0.1553 | 0.4378 | |||
| TAAG | 0.443 | 0.460, 0.435 | 0.4589 | 0.9270 | |||
| TAGC | 0.027 | 0.027, 0.027 | 0.9495 | >0.9999 | |||
Table┬Ā5.
| Phenotype | Best interaction model | Testing accuracy (%) | CVC | Adjusted p |
|---|---|---|---|---|
| ADHD-male | ESR2 rs960070/BDNF rs6265/BDNF rs2049046/SNAP25 rs362987/CDH13 rs6565113 | 54.57 | 8/10 | 0.0004*** |
| ADHD-female | ESR2 rs960070/BDNF rs6265/BDNF rs2049046 | 63.82 | 8/10 | <0.0002*** |
Table┬Ā6.
REFERENCES







