|
 |
- Search
| Psychiatry Investig > Volume 8(2); 2011 > Article |
Abstract
Objective
Psychiatric disorders such as depression, anxiety and alcohol dependence are associated with serotonin metabolism. We assessed the methylation level of the serotonin transporter (5-HTT) promoter region in control and alcohol dependent patients.
Methods
Twenty seven male patients who met the Diagnostic and Statistical Manual of Mental Disorder IV (DSM-IV) criteria for alcohol dependence were compared with fifteen controls. Polymerase chain reaction (PCR) assays of bisulfate-modified DNA were designed to amplify a part of the CpG island in the 5HTT gene. Pyrosequencing was performed and the methylation level at seven CpG island sites was measured.
Results
We found no differences in the methylation patterns of the serotonin transporter linked promoter region (5-HTTLPR) between
alcohol-dependent and control subjects.
Conclusion
Our negative finding may be because 5-HTT epigenetic variation may not affect the expression for 5-HTT or there may be other methylation site critical for its expression. To find out more conclusive result, repeating the study in more methylation sites with a larger number of samples in a well-controlled setting is needed.
Psychiatric disorders such as major depression, obsessive compulsive disorder, autism, and alcohol dependence are associated with serotonin neurotransmission.1-4 The serotonin transporter (5-HTT) is an integral membrane protein that transports the neurotransmitter serotonin from the synaptic space into presynaptic neurons and is a member of the SLC6 gene family, which reuptakes other neurotransmitters (e.g., glycine, gamma-aminobutyric acid, dopamine, and norepinephrine) across the plasma membrane of neurons and glia.5 There are many studies regarding 5-HTT genetic polymorphisms and the subsequent affects on etiology and response to treatment in mental illness.6-9
Feinn et al.4 reported a significant association with the frequency of the short (S) allele at the 5-HTT linked promoter region (5-HTTLPR) in patients with alcohol dependence. In a meta-analysis, the S allele was significantly associated with an early onset, severe subtype and a co-morbid psychiatric disorder in alcohol dependence. Decreased raphe 5-HTT, which correlated with lifetime alcohol consumption, and association of the 5-HTT gene regulatory polymorphism in patients with alcohol dependence were reported.10 Hinckers et al.11 reported that carriers of two long (L) alleles of the 5-HTT regulatory region had a lower response to alcohol than carriers of one or two S alleles among adolescents.
The CpG island that surrounds exon 1 was recently discovered; its role in psychiatric disorders is unknown. Effects of 5-HTTLPR on 5-HTT mRNA production is modest and other cis- and trans-acting elements involved in the regulation of the 5-HTT gene region, such as the CpG island, constitute the leading mechanisms that control 5-HTT activity.12 Although heritable components affect gene activity, gene-environment interactions may contribute to gene status by gene regulatory elements such as CpG islands.
However Philibert et al.16 reported relation between the methylation level of 5-HTT promoter region and well known 5-HTTLPR polymorphism. They opened the possibility that 5-HTTLPR polymorphism may be the reflection of methylation difference of 5-HTT promoter region.
We hypothesized that there was a relationship between serotonin transporter epigenetic changes and alcohol dependence; thus, we investigated the differences in quantitative CpG residue methylation of seven 5-HTT promoter CpG positions between control subjects and patients with alcohol dependence.
We assessed the 5-HTT promoter methylation level of the subjects. Twenty seven male patients who met the Diagnostic and Statistical Manual of Mental Disorder IV (DSM-IV)13 criteria for alcohol dependence were interviewed and sampled. They were all admitted to Hangang Sacred Heart Hospital. Two hospital psychiatrists determined the diagnosis in a consensus procedure using all available clinical material, including a semistructured interview based on the DSM-IV. Patients were excluded from the study if they had a history of comorbid psychiatric illness or physical disorders other than alcohol-related medical illnesses. The control subjects consisted of fifteen unrelated healthy males, mostly nondrinkers. A few of the control subjects were occasional light drinkers, as revealed by a drinking-habit questionnaire.
Mean age for the control subjects and alcohol dependent patients was 44.93±6.47 and 46.67±13.74 years, respectively. The two groups did not differ significantly in age (t=-0.46, df= 17.5, p=0.65). The institutional review board of Hangang Sacred Heart Hospital approved the study and all subjects provided informed consent after a complete description of the present study.
Alcohol dependent patients consumed alcohol almost continually before admission. Ten mL of fasting blood was drawn into an EDTA tube on the day after admission and the blood was immediately centrifuged at 3,800 rpm for 10 minutes. Plasma was stored at -70℃ until it was thawed for assay.
Discrimination of methylated and unmethylated site of CpG island were done Bisulfate modification. Bisulfite treatment changes unmethylated cytosine residues to uracil, but leaves methylated 5-methylcytosine unaffected. Bisulfite treatment of 200 ng of each sample was accomplished using the EZ DNA Methylation-Gold kit (Zymo Research, Orange, CA, USA) according to the manufacturer's instructions. The bisulfite converted DNA was eluted in 20 uL of elution buffer (Zymo Research, Orange, CA, USA). DNA samples were immediately stored at -20℃ until use.
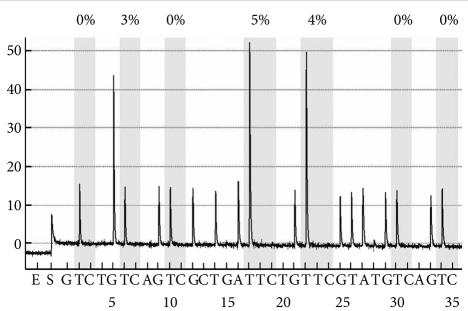
Pyrosequencing were used to identify sequence change after bisulfate modification. Pyrogram (Figure 1) shows DNA sequence representation with sequential addition of dNTP detected by luciferase in one sample. Methylation Index were described above gray area which represents each CpG sites.
Polymerase chain reaction (PCR) assays were designed to amplify a part of the CpG island in the 5HTT gene. Primers were designed using the PSQ assay design program (Biotage, Charlotte, NC, USA). The primer sequences are listed in Table 1. The PCR reaction was carried out in a volume of 50 uL with 20 ng or less converted gDNA, 5 uL of 10X Taq buffer, 5 units Hot Start Taq polymerase (Enzynomics, Korea), 4 uL of 2.5mM dNTP mixture, and 2 uL of 10 pmole/uL PCR primers. Amplification was carried out according to the general guidelines suggested by pyrosequencing: denaturating at 95℃ for 5 min, followed by 45 cycles at 95℃ for 40 sec, 60℃ for 45 sec, and 72℃ for 40 sec and a final extension at 72℃ for 5 min. Confirmation of PCR product quality was established on a 1.5% agarose gel stained with ethidium bromide. Pyrosequencing was performed using the PSQ96MA System (Biotage, Charlotte, NC, USA) according to the manufacturer's instructions, including single strand binding protein (Pyro Gold reagents; Qiagen Inc., Valencia, CA, USA). PCR primer sequences, PCR conditions and sequencing primer sequences are given in Table 1.
The methylation index (MtI) at the promoter and for the samples was calculated as the average value of methylated cytosine/(methylated cytosine+unmethylated cytosine) for all examined CpGs in the gene. All data were analyzed using SPSS for Windows software (version 12.0; SPSS Inc., Chicago, IL, USA). All analyses were set at a two-tailed significance level of p<0.05.
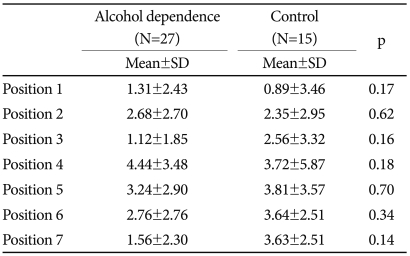
An example of a bisulfite methylation profile is shown in Figure 1.There was no difference in MtI between patients with alcohol dependence and controls at any of the seven CpG residues (Table 2). We considered low-level methylation (0-5%) to represent background 'noise' with questionable significance.
Complex epigenetic mechanisms that regulate gene activity without altering DNA code produce long-lasting changes in gene expression essential to development, cellular differentiation, and to adaptation to environmental changes. These mechanisms that include DNA methylation, post-translational covalent modifications of histones, nucleosome sliding and nucleosome and histone substitution, cause modifications of chromatin conformation, which, in turn, reglate gene expression.14 For example, the amount of DNA methylation in promoter regions correlates with gene inactivation. Currently, with regard to alcohol dependence, alpha synuclein (SNCA), alcohol dehydrogenase and HTT genes are often studied in epigenetics. This study examined serotonin transporter epigenetic changes in alcohol dependent patients. However, we found no differences in the methylation patterns of the serotonin transporter promoter region between alcohol dependent and control subjects.
Currently, with regard to alcohol intoxication, there are few studies regarding methylation patterns of the serotonin transporter promoter. Philibert et al.15 found that females had more CpG methylation in the 5-HTT gene and less mRNA expression. They discovered a trend of increased overall methylation in people with a lifetime history of major depression. However, there was higher 5-HTT mRNA expression in patients with a lifetime history of alcohol dependence.16
Serotonin transporter mRNA expression is affected by 5-HTTLPR genotype.17 However, there was no relationship with overall CpG methylation level in the present study. This result is in accordance with Philbert et al.,15 who didn't found the main effect of total methylation and 5-HTT mRNA level. Although increased average methylation only after 5HTTLPR genotype (l/l or s/s) was taken into account; effects of CpG methylation were then evident.
The epigenetics field and its methodologies are relatively new and limited.18 To identify epigenetic changes associated with alcohol dependence, the ideal experiment would prospectively investigate the dynamics of genome-wide epigenetic changes in the brains of individuals who eventually become affected with alcohol dependence, as compared to unaffected control individuals. At the present time, however, limitations with our current range of epigenetic methodologies render such an approach impossible. In addition, because there have been no thorough epigenetic studies with regard to alcohol dependence to date, it is not known how large a sample would need to be investigated. At this stage, the degree of any potential epigenetic differences between cases and controls is unknown, although the effects observed are likely to be small.
This study has an important limitation. Peripheral blood-based assays will progress after properly controlling different confounding variables that have an effect on DNA methylation, including medications, plasma homocysteine level,19 and underlying disease.20 However, the use of peripheral cells would also allow the longitudinal study of epigenetic changes throughout the life course of patients with alcohol dependence, providing information about the epigenetic factors associated with the development, remission and relapse of the disorder.18
Conclusively, in this research, there were no differences of 5-HTTLPR epigenetic DNA methylation between patients with alcohol dependence and control subjects. It is thought that 5-HTT epigenetic variation may not affect the expression for 5-HTT or there may be other methylation site critical for its expression.
Acknowledgments
This study was supported by a grant from the Korean Neuropsychiatric Association, Republic of Korea.
References
1. Lin PY. Meta-analysis of the association of serotonin transporter gene polymorphism with obsessive-compulsive disorder. Prog Neuropsychopharmacol Biol Psychiatry 2007;31:683-689. PMID: 17291658.


2. Brown GW, Harris TO. Depression and the serotonin transporter 5-HTTLPR polymorphism: a review and a hypothesis concerning gene-environment interaction. J Affect Disord 2008;111:1-12. PMID: 18534686.


3. Huang CH, Santangelo SL. Autism and serotonin transporter gene polymorphisms: a systematic review and meta-analysis. Am J Med Genet B Neuropsychiatr Genet 2008;147B:903-913. PMID: 18286633.


4. Feinn R, Nellissery M, Kranzler HR. Meta-analysis of the association of a functional serotonin transporter promoter polymorphism with alcohol dependence. Am J Med Genet B Neuropsychiatr Genet 2005;133B:79-84. PMID: 15635638.


5. Hahn MK, Blakely RD. The functional impact of SLC6 transporter genetic variation. Annu Rev Pharmacol Toxicol 2007;47:401-441. PMID: 17067279.


6. Conroy J, Meally E, Kearney G, Fitzgerald M, Gill M, Gallagher L. Serotonin transporter gene and autism: a haplotype analysis in an Irish autistic population. Mol Psychiatry 2004;9:587-593. PMID: 14708029.


7. Gorwood P. Eating disorders, serotonin transporter polymorphisms and potential treatment response. Am J Pharmacogenomics 2004;4:9-17. PMID: 14987118.


8. Lotrich FE, Pollock BG. Meta-analysis of serotonin transporter polymorphisms and affective disorders. Psychiatr Genet 2004;14:121-129. PMID: 15318024.


9. Savitz JB, Ramesar RS. Genetic variants implicated in personality: a review of the more promising candidates. Am J Med Genet B Neuropsychiatr Genet 2004;131B:20-32. PMID: 15389772.


10. Heinz A, Ragan P, Jones DW, Hommer D, Williams W, Knable MB, et al. Reduced central serotonin transporters in alcoholism. Am J Psychiatry 1998;155:1544-1549. PMID: 9812115.


11. Hinckers AS, Laucht M, Schmidt MH, Mann KF, Schumann G, Schuckit MA, et al. Low level of response to alcohol as associated with serotonin transporter genotype and high alcohol intake in adolescents. Biol Psychiatry 2006;60:282-287. PMID: 16497275.


12. Bradley SL, Dodelzon K, Sandhu HK, Philibert RA. Relationship of serotonin transporter gene polymorphisms and haplotypes to mRNA transcription. Am J Med Genet B Neuropsychiatr Genet 2005;136B:58-61. PMID: 15858822.


13. American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorder, Fourth Edition. 1994,Washington, DC: American Psychiatric Press.
14. Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci 2007;8:355-367. PMID: 17453016.


15. Philibert RA, Sandhu H, Hollenbeck N, Gunter T, Adams W, Madan A. The relationship of 5HTT (SLC6A4) methylation and genotype on mRNA expression and liability to major depression and alcohol dependence in subjects from the Iowa Adoption Studies. Am J Med Genet B Neuropsychiatr Genet 2008;147B:543-549. PMID: 17987668.


16. Philibert R, Madan A, Andersen A, Cadoret R, Packer H, Sandhu H. Serotonin transporter mRNA levels are associated with the methylation of an upstream CpG island. Am J Med Genet B Neuropsychiatr Genet 2007;144B:101-105. PMID: 16958039.


17. Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, et al. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science 1996;274:1527-1531. PMID: 8929413.


18. Mill J, Petronis A. Molecular studies of major depressive disorder: the epigenetic perspective. Mol Psychiatry 2007;12:799-814. PMID: 17420765.


19. Yi P, Melnyk S, Pogribna M, Pogribny IP, Hine RJ, James SJ. Increase in plasma homocysteine associated with parallel increases in plasma S-adenosylhomocysteine and lymphocyte DNA hypomethylation. J Biol Chem 2000;275:29318-29323. PMID: 10884384.


20. Lenz B, Soehngen C, Linnebank M, Heberlein A, Frieling H, Kornhuber J, et al. Genetic polymorphisms relevant for one-carbon metabolism show no effect on homocysteine plasma levels and DNA methylation in alcoholism. Psychiatr Genet 2009;19:215-216. PMID: 19461557.