 |
 |
- Search
| Psychiatry Investig > Volume 20(12); 2023 > Article |
|
Abstract
Objective
Methods
Results
Notes
Availability of Data and Material
All data generated or analyzed during the study are included in this published article.
Conflicts of Interest
The authors have no potential conflicts of interest to disclose.
Author Contributions
Conceptualization: Zhi-Lin Luan. Data curation: Yang Sun, Zhi-Lin Luan. Formal analysis: Yang Sun, Lin Wen. Funding acquisition: Zhi-Lin Luan. Methodology: Lin Wen, Yi-Yang Luo, Wen-Juan Hu, Cong Zhang, Ping Gao, Li-Na Xuan. Project administration: Zhi-Lin Luan. Resources: Yang Sun. Software: Ye Lv. Supervision: Zhi-Lin Luan. Validation: Guan-Yu Wang, Cheng-Jie Li, Zhi-Xin Xiang. Visualization: Yang Sun, Lin Wen. WritingŌĆöoriginal draft: Yi-Yang Luo. WritingŌĆöreview & editing: Zhi-Lin Luan, Hui-Wen Ren.
Funding Statement
This work was supported by Natural Science Foundation of Liaoning Province, China 2022-MS-326 (to Z.L.), Education Department of Liaoning Province, China LZ2020023 (to Z.L.), and the Dalian Young Star of Science and Technology 2019RQ116 (to Z.L.). We are also grateful for the support from Liaoning BaiQianWan Talents Program.
Figure┬Ā1.

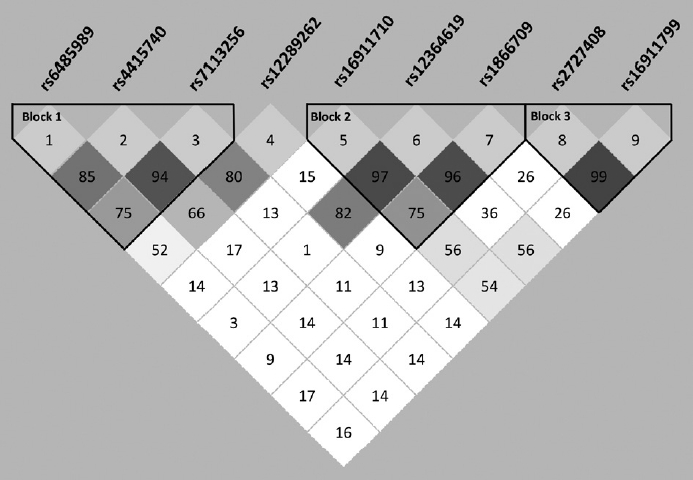
Figure┬Ā2.
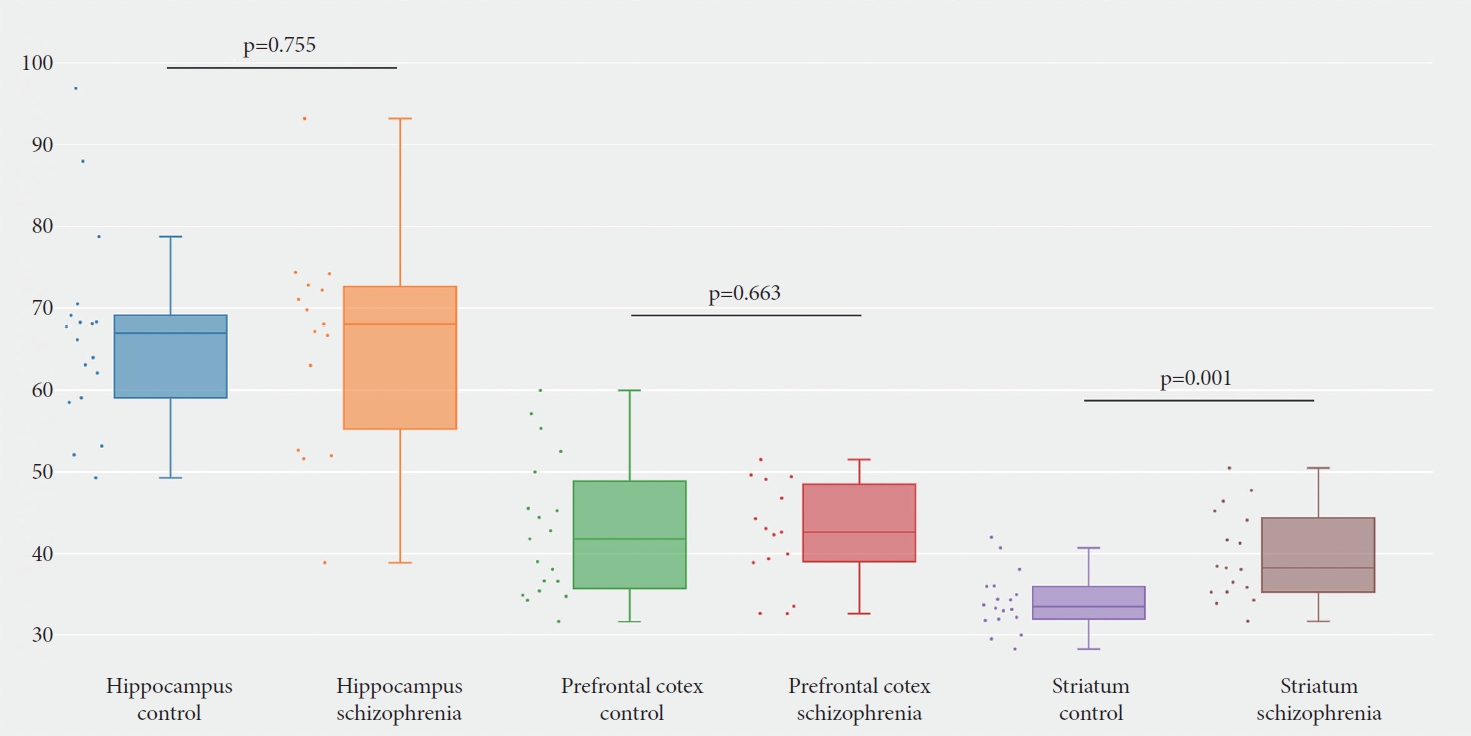
Figure┬Ā3.
Table┬Ā1.
| Case (N=721) | Control (N=1,195) | |
|---|---|---|
| Age (yr) | 28.808┬▒9.900 | 27.874┬▒9.510 |
| Sex | ||
| ŌĆāMale | 334 | 603 |
| ŌĆāFemale | 387 | 592 |
Table┬Ā2.
| No. | rs code | Position | Distance from SNP1 (kb) | Location | Allele change | HCB* MAF | Sample-set HWE p | Case HWE p | Control HWE p |
|---|---|---|---|---|---|---|---|---|---|
| SNP1 | rs6485989 | 12673624 | 0 | 5' near gene | C>T | 0.395 | 0.145 | 0.176 | 0.362 |
| SNP2 | rs4415740 | 12762364 | 89 | Intron | C>T | 0.395 | 0.312 | 0.102 | 0.806 |
| SNP3 | rs7113256 | 12824302 | 151 | Intron | C>T | 0.209 | 0.319 | 0.110 | 0.770 |
| SNP4 | rs12289262 | 12873211 | 200 | Intron | C>T | 0.326 | 0.067 | 0.188 | 0.167 |
| SNP5 | rs16911710 | 12889474 | 216 | Intron | G>T | 0.467 | 0.157 | 0.474 | 0.222 |
| SNP6 | rs12364619 | 12898724 | 225 | Intron | C>T | 0.010 | 0.678 | 0.734 | 0.793 |
| SNP7 | rs1866709 | 12913904 | 240 | Intron | G>A | 0.302 | 0.985 | 0.897 | 0.985 |
| SNP8 | rs2727408 | 12920305 | 247 | Intron | A>C | 0.302 | 0.346 | 0.373 | 0.613 |
| SNP9 | rs16911799 | 12929006 | 255 | Intron | G>A | 0.163 | 0.484 | 0.427 | 0.781 |
* Chinese Han population MAF from the International HapMap Project Database (http://www.ncbi.nlm.nih.gov/SNP/).
Table┬Ā3.
| No. | rs code | Subjects | Allele and frequencyŌĆĪ | Žć2, p (pŌĆĀ) | Genotype and frequencyŌĆĪ | Žć2, p (pŌĆĀ) | OR (95% CI) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| C | T | CC | CT | TT | ||||||
| SNP1 | rs6485989 | Case | 1,039 (0.721) | 403 (0.279) | Žć2=6.313 | 367 (0.509) | 305 (0.423) | 49 (0.068) | Žć2=6.799 | 1.202 (1.041-1.388) |
| Control | 1,630 (0.682) | 760 (0.318) | p=0.012* | 549 (0.459) | 532 (0.445) | 114 (0.095) | p=0.034* | |||
| C | T | CC | CT | TT | ||||||
| SNP2 | rs4415740 | Case | 1,037 (0.719) | 405 (0.281) | Žć2=11.564 | 364 (0.505) | 309 (0.429) | 48 (0.067) | Žć2=13.024 | 1.281 (1.110-1.478) |
| Control | 1,593 (0.667) | 797 (0.333) | p=0.001* (0.006)* | 529 (0.443) | 535 (0.448) | 131 (0.110) | p=0.002* (0.014)* | |||
| C | T | CC | CT | TT | ||||||
| SNP3 | rs7113256 | Case | 1,159 (0.804) | 283 (0.196) | Žć2=11.493 | 459 (0.637) | 241 (0.334) | 21 (0.029) | Žć2=12.867 | 1.318 (1.123-1.547) |
| Control | 1,808 (0.756) | 582 (0.244) | p=0.001* (0.006)* | 682 (0.571) | 444 (0.372) | 69 (0.058) | p=0.002* (0.015)* | |||
| C | T | CC | CT | TT | ||||||
| SNP4 | rs12289262 | Case | 1,108 (0.769) | 332 (0.231) | Žć2=4.838 | 420 (0.583) | 268 (0.372) | 32 (0.044) | Žć2=5.110 | 1.187 (1.019-1.383) |
| Control | 1,763 (0.738) | 627 (0.262) | p=0.028* | 641 (0.536) | 481 (0.403) | 73 (0.061) | p=0.078 | |||
| G | T | GG | GT | TT | ||||||
| SNP5 | rs16911710 | Case | 1,045 (0.726) | 395 (0.274) | Žć2=1.367 | 383 (0.532) | 279 (0.388) | 58 (0.081) | Žć2=1.365 | 0.916 (0.790-1.061) |
| Control | 1,774 (0.743) | 614 (0.257) | p=0.242 | 667 (0.559) | 440 (0.369) | 87 (0.073) | p=0.505 | |||
| C | T | CC | CT | TT | ||||||
| SNP6 | rs12364619 | Case | 18 (0.013) | 1,422 (0.988) | Žć2=2.382 | 0 | 18 (0.025) | 702 (0.975) | Žć2=2.405 | 1.668 (0.865-3.217) |
| Control | 18 (0.008) | 2,372 (0.992) | p=0.123 | 0 | 18 (0.015) | 1,177 (0.985) | p=0.121 | |||
| A | G | AA | AG | GG | ||||||
| SNP7 | rs1866709 | Case | 249 (0.173) | 1,193 (0.827) | Žć2=8.185 | 21 (0.029) | 207 (0.287) | 493 (0.684) | Žć2=8.185 | 1.298 (1.085-1.553) |
| Control | 331 (0.138) | 2,059 (0.862) | p=0.004* (0.018)* | 23 (0.019) | 285 (0.238) | 887 (0.742) | p=0.017* | |||
| A | C | AA | AC | CC | ||||||
| SNP8 | rs2727408 | Case | 800 (0.555) | 641 (0.445) | Žć2=0.508 | 216 (0.300) | 368 (0.510) | 137 (0.190) | Žć2=0.671 | 0.953 (0.836-1.087) |
| Control | 1,353 (0.567) | 1,035 (0.433) | p=0.476 | 379 (0.317) | 595 (0.495) | 220 (0.184) | p=0.715 | |||
| A | G | AA | AG | GG | ||||||
| SNP9 | rs16911799 | Case | 631 (0.438) | 809 (0.562) | Žć2=0.860 | 133 (0.185) | 365 (0.507) | 222 (0.308) | Žć2=1.076 | 1.064 (0.933-1.215) |
| Control | 1,009 (0.423) | 1,377 (0.577) | p=0.354 | 211 (0.177) | 587 (0.492) | 395 (0.331) | p=0.584 | |||
Table┬Ā4.
| Block | Haplotype |
Haplotype frequencyŌĆĀ |
Žć2 | p | OR (95% CI) |
Global |
||
|---|---|---|---|---|---|---|---|---|
| Case | Control | Žć2 | p | |||||
| SNP1-SNP2-SNP3 | C-C-C | 973.57 (0.675) | 1,510.64 (0.632) | 7.328 | 0.007* | 1.214 (1.055-1.396) | 15.152 | 0.004* |
| C-T-T | 43.77 (0.030) | 82.41 (0.034) | 0.488 | 0.485 | 0.876 (0.603-1.271) | |||
| T-C-C | 47.78 (0.033) | 67.93 (0.028) | 0.675 | 0.411 | 1.171 (0.804-1.705) | |||
| T-T-C | 131.62 (0.091) | 205.72 (0.086) | 0.294 | 0.588 | 1.065 (0.847-1.340) | |||
| T-T-T | 223.59 (0.155) | 485.16 (0.203) | 13.860 | <0.001* | 0.719 (0.604-0.856) | |||
| SNP5-SNP6-SNP7 | G-T-A | 229.82 (0.160) | 310.96 (0.130) | 7.148 | 0.008* | 1.287 (1.069-1.548) | 10.257 | 0.006* |
| G-T-G | 795.19 (0.553) | 1,445.05 (0.605) | 8.195 | 0.004* | 0.822 (0.718-0.940) | |||
| T-T-G | 376.81 (0.262) | 595.04 (0.249) | 1.128 | 0.288 | 1.085 (0.933-1.261) | |||
| SNP8-SNP9 | A-G | 794.97 (0.552) | 1,343.92 (0.564) | 0.729 | 0.393 | 0.944 (0.826-1.078) | 0.729 | 0.393 |
| C-A | 627.97 (0.436) | 1,001.92 (0.420) | 0.729 | 0.393 | 1.060 (0.928-1.210) | |||
REFERENCES
-
METRICS

-
- 0 Crossref
- Scopus
- 1,307 View
- 29 Download
-
Association of
CACNA1C Variants with Bipolar Disorder in the Korean Population2016 July;13(4)






